Exploring the comparative genome of rice pathogen Burkholderia plantarii: unveiling virulence, fitness traits, and a potential type III secretion system effector
- PMID: 38845849
- PMCID: PMC11153758
- DOI: 10.3389/fpls.2024.1416253
Exploring the comparative genome of rice pathogen Burkholderia plantarii: unveiling virulence, fitness traits, and a potential type III secretion system effector
Abstract
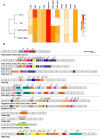
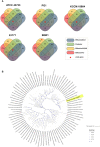
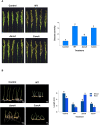
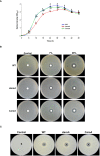
This study presents a comprehensive genomic analysis of Burkholderia plantarii, a rice pathogen that causes blight and grain rot in seedlings. The entire genome of B. plantarii KACC 18964 was sequenced, followed by a comparative genomic analysis with other available genomes to gain insights into its virulence, fitness, and interactions with rice. Multiple secondary metabolite gene clusters were identified. Among these, 12 demonstrated varying similarity levels to known clusters linked to bioactive compounds, whereas eight exhibited no similarity, indicating B. plantarii as a source of potentially novel secondary metabolites. Notably, the genes responsible for tropolone and quorum sensing were conserved across the examined genomes. Additionally, B. plantarii was observed to possess three complete CRISPR systems and a range of secretion systems, exhibiting minor variations among the analyzed genomes. Genomic islands were analyzed across the four genomes, and a detailed study of the B. plantarii KACC 18964 genome revealed 59 unique islands. These islands were thoroughly investigated for their gene contents and potential roles in virulence. Particular attention has been devoted to the Type III secretion system (T3SS), a crucial virulence factor. An in silico analysis of potential T3SS effectors identified a conserved gene, aroA. Further mutational studies, in planta and in vitro analyses validated the association between aroA and virulence in rice. Overall, this study enriches our understanding of the genomic basis of B. plantarii pathogenicity and emphasizes the potential role of aroA in virulence. This understanding may guide the development of effective disease management strategies.
Keywords: genome analysis; rice pathogenic bacteria; secondary metabolites; type III secretion system; virulence.
Copyright © 2024 Mannaa, Lee, Lee, Han, Kang, Kim, Park and Seo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures



Similar articles
-
Impact of Quorum Sensing on the Virulence and Survival Traits of Burkholderia plantarii.Plants (Basel). 2024 Sep 23;13(18):2657. doi: 10.3390/plants13182657. Plants (Basel). 2024. PMID: 39339632 Free PMC article.
-
Comparative genome analysis of rice-pathogenic Burkholderia provides insight into capacity to adapt to different environments and hosts.BMC Genomics. 2015 May 6;16(1):349. doi: 10.1186/s12864-015-1558-5. BMC Genomics. 2015. PMID: 25943361 Free PMC article.
-
Repression of tropolone production and induction of a Burkholderia plantarii pseudo-biofilm by carot-4-en-9,10-diol, a cell-to-cell signaling disrupter produced by Trichoderma virens.PLoS One. 2013 Nov 4;8(11):e78024. doi: 10.1371/journal.pone.0078024. eCollection 2013. PLoS One. 2013. PMID: 24223754 Free PMC article.
-
Functional and genomic insights into the pathogenesis of Burkholderia species to rice.Environ Microbiol. 2016 Mar;18(3):780-90. doi: 10.1111/1462-2920.13189. Epub 2016 Jan 21. Environ Microbiol. 2016. PMID: 26690879 Review.
-
Burkholderia glumae: next major pathogen of rice?Mol Plant Pathol. 2011 May;12(4):329-39. doi: 10.1111/j.1364-3703.2010.00676.x. Epub 2010 Nov 24. Mol Plant Pathol. 2011. PMID: 21453428 Free PMC article. Review.
Cited by
-
Impact of Quorum Sensing on the Virulence and Survival Traits of Burkholderia plantarii.Plants (Basel). 2024 Sep 23;13(18):2657. doi: 10.3390/plants13182657. Plants (Basel). 2024. PMID: 39339632 Free PMC article.
References
-
- Azegami K., Nishiyama K., Watanabe Y., Kadota I., Ohuchi A., Fukazawa C. (1987). Pseudomonas plantarii sp. nov., the causal agent of rice seedling blight. Int. J. Syst. Evol. Microbiol. 37, 144–152. doi: 10.1099/00207713-37-2-144 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

