Deciphering genetic diversity phylogeny and assembly of Allium species through micro satellite markers on nuclear DNA
- PMID: 38845887
- PMCID: PMC11153109
- DOI: 10.1016/j.heliyon.2024.e31650
Deciphering genetic diversity phylogeny and assembly of Allium species through micro satellite markers on nuclear DNA
Abstract
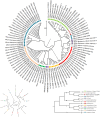
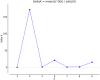
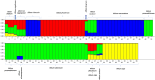
The genus Allium is the most diverse, with cultivated crops such as onion, garlic, bunching onion, chives, leeks, and shallots, and several wild and semi-domesticated Allium species utilized as minor vegetables. These minor species are the genetic resources for various abiotic and biotic stresses. To employ underutilized species in breeding programmes, the magnitude of the genetic background of cultivated and semi-domesticated alliums, the phylogeny and diversity of the population must be known. In this study, nineteen SSR markers were employed to study the divergence and population structure of 95 Allium accessions which includes species, varieties, and interspecific hybrids, yielded 92 polymorphic loci, averaging 4.84 loci per SSR. PIC values range between 0.24 (ACM 018) and 0.98 (ACM 099). The cross transferability of ACM markers among Allium species ranges from 1.33 to 10.53 per cent, which is relatively low. The genotypes investigated were clustered into four primary clusters A, B, C, and D with 13 sub clusters I-XIII, conferring to the clustering results. The population structure investigations also found that K is a peak at value 4, implying that the population is predominantly segregated into four distinct groups, which associates the clustering pattern. The employed SSR markers adeptly unravel the complexities of diversity within alliums, holding promise for refining future breeding programs targeting elite progenies.
Keywords: Allium; Diversity; Molecular markers; Population structure; SSR; Wild species.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Microsatellite analysis and marker development in garlic: distribution in EST sequence, genetic diversity analysis, and marker transferability across Alliaceae.Mol Genet Genomics. 2018 Oct;293(5):1091-1106. doi: 10.1007/s00438-018-1442-5. Epub 2018 Apr 28. Mol Genet Genomics. 2018. PMID: 29705936
-
SSR markers development and their application in genetic diversity evaluation of garlic (Allium sativum) germplasm.Plant Divers. 2021 Aug 11;44(5):481-491. doi: 10.1016/j.pld.2021.08.001. eCollection 2022 Sep. Plant Divers. 2021. PMID: 36187554 Free PMC article.
-
Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome.BMC Plant Biol. 2004 Jul 14;4:11. doi: 10.1186/1471-2229-4-11. BMC Plant Biol. 2004. PMID: 15253775 Free PMC article.
-
Allium vegetables for possible future of cancer treatment.Phytother Res. 2019 Dec;33(12):3019-3039. doi: 10.1002/ptr.6490. Epub 2019 Aug 29. Phytother Res. 2019. PMID: 31464060 Review.
-
Allium vegetables in cancer prevention: an overview.Asian Pac J Cancer Prev. 2004 Jul-Sep;5(3):237-45. Asian Pac J Cancer Prev. 2004. PMID: 15373701 Review.
References
-
- Benke A.P., Mahajan V., Manjunathagowda D.C., Mokat D.N. Interspecific hybridization in Allium crops: status and prospectus. Genet. Resour. Crop Evol. 2022;69:1–9. doi: 10.1007/s10722-021-01283-5. - DOI
-
- ITIS Interagency taxonomic Information system. 2021. https://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search...
-
- Vavilov N.I. Centers of origin of cultivated plants. Bull Appl Bot Plant Breed. 1926;16:1–248. https://ci.nii.ac.jp/naid/10016876337/
-
- Pandey A., Pandey R., Negi K.S., Radhamani J. Realizing value of genetic resources of Allium in India. Genet. Resour. Crop Evol. 2008;55:985–994. doi: 10.1007/s10722-008-9305-2. - DOI
-
- Manjunathagowda D.C. Perspective and application of molecular markers linked to the cytoplasm types and male-fertility restorer locus in onion (Allium cepa) Plant Breed. 2021 doi: 10.1111/PBR.12948. pbr. - DOI
LinkOut - more resources
Full Text Sources

