CircRNA hsa_circ_0069,399 as a potential clinical prognostic marker in laryngeal squamous cell carcinoma
- PMID: 38845955
- PMCID: PMC11154600
- DOI: 10.1016/j.heliyon.2024.e31969
CircRNA hsa_circ_0069,399 as a potential clinical prognostic marker in laryngeal squamous cell carcinoma
Erratum in
-
Corrigendum to "The effect of caffeine supplementation on muscular strength and endurance: A meta-analysis of meta-analyses" [Heliyon 10(15), August 2024, e35025].Heliyon. 2024 Nov 21;10(23):e40064. doi: 10.1016/j.heliyon.2024.e40064. eCollection 2024 Dec 15. Heliyon. 2024. PMID: 39759852 Free PMC article.
Abstract
Objective: Circular RNAs (circRNAs) significantly influence the invasion, metastasis, gene expression, proliferation, and apoptosis of tumor cells. However, the roles of circRNAs in laryngeal squamous cell carcinoma (LSCC) remain largely unexplored. This study aims to examine circRNA expression patterns in LSCC and adjacent non-tumorous tissues, with the goal of uncovering potential biomarkers for LSCC.
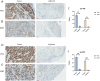
Methods: Tissue samples were collected from both the tumor and adjacent normal tissues of ten patients who had undergone surgical resection. The profiling of circRNAs was conducted through transcriptomic sequencing and analytical bioinformatics approaches. A ternary regulatory network based on the competitive endogenous RNA (ceRNA) hypothesis was established, linking target circRNAs to clinical immunohistochemical parameters for comparison. Verification of target circRNAs in LSCC tissues was performed using quantitative real-time PCR (RT-qPCR), whereas target mRNAs were analyzed through immunohistochemistry.
Results: A total of 126 significantly different circRNAs were identified, including 40 up-regulated genes and 86 down-regulated genes. Furthermore, 92 circRNA-miRNA-mRNA regulatory relationship axes related to clinical immunohistochemical indicators were found based on 5 candidate circRNAs. Interestingly, all axes related to the target genes MKI67 and TP53 were found to compete with the same circRNA: hsa_circ_0069,399. Further verification confirmed that the hsa_circ_0069,399 expression was overtly upregulated in tumor tissues from LSCC patients, which was consistent with the sequencing results.
Conclusion: hsa_circ_0069,399 could be a potential prognostic marker for LSCC.
Keywords: Laryngeal squamous cell carcinoma; ceRNA; circRNA; hsa_circ_0069399.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Schutte H.W., Heutink F., Wellenstein D.J., et al. Impact of time to diagnosis and treatment in head and neck cancer: a systematic review. Otolaryngol. Head Neck Surg. 2020;162(4):446–457. - PubMed
-
- Pan X.L., Lin Y. The correct selection of treatment strategies for laryngeal cancer to improve the survival rate and quality of life for patients. Zhonghua er bi yan hou tou jing wai ke za zhi. 2020;55(12):1111–1115. - PubMed
-
- Zheng Z.X.G. Treatment strategy and progress of laryngeal cancer. Modern oncology. 2022;30(11):2090–2093.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

