LCASPMDA: a computational model for predicting potential microbe-drug associations based on learnable graph convolutional attention networks and self-paced iterative sampling ensemble
- PMID: 38846568
- PMCID: PMC11153849
- DOI: 10.3389/fmicb.2024.1366272
LCASPMDA: a computational model for predicting potential microbe-drug associations based on learnable graph convolutional attention networks and self-paced iterative sampling ensemble
Abstract
Introduction: Numerous studies show that microbes in the human body are very closely linked to the human host and can affect the human host by modulating the efficacy and toxicity of drugs. However, discovering potential microbe-drug associations through traditional wet labs is expensive and time-consuming, hence, it is important and necessary to develop effective computational models to detect possible microbe-drug associations.
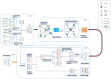
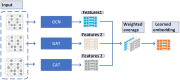
Methods: In this manuscript, we proposed a new prediction model named LCASPMDA by combining the learnable graph convolutional attention network and the self-paced iterative sampling ensemble strategy to infer latent microbe-drug associations. In LCASPMDA, we first constructed a heterogeneous network based on newly downloaded known microbe-drug associations. Then, we adopted the learnable graph convolutional attention network to learn the hidden features of nodes in the heterogeneous network. After that, we utilized the self-paced iterative sampling ensemble strategy to select the most informative negative samples to train the Multi-Layer Perceptron classifier and put the newly-extracted hidden features into the trained MLP classifier to infer possible microbe-drug associations.
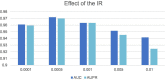
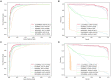
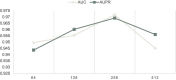
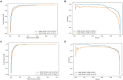
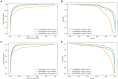
Results and discussion: Intensive experimental results on two different public databases including the MDAD and the aBiofilm showed that LCASPMDA could achieve better performance than state-of-the-art baseline methods in microbe-drug association prediction.
Keywords: drug-microbe association; learnable graph convolutional attention network; multi-layer perceptron classifier; prediction model; self-paced iterative sampling ensemble.
Copyright © 2024 Yang, Wang, Zhang, Zeng, Zhang and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Predicting microbe-drug associations with structure-enhanced contrastive learning and self-paced negative sampling strategy.Brief Bioinform. 2023 Mar 19;24(2):bbac634. doi: 10.1093/bib/bbac634. Brief Bioinform. 2023. PMID: 36715986
-
Prediction of Human Microbe-Drug Association based on Layer Attention Graph Convolutional Network.Curr Med Chem. 2024;31(31):5097-5109. doi: 10.2174/0109298673249941231108091326. Curr Med Chem. 2024. PMID: 39225188
-
GSAMDA: a computational model for predicting potential microbe-drug associations based on graph attention network and sparse autoencoder.BMC Bioinformatics. 2022 Nov 18;23(1):492. doi: 10.1186/s12859-022-05053-7. BMC Bioinformatics. 2022. PMID: 36401174 Free PMC article.
-
GACNNMDA: a computational model for predicting potential human microbe-drug associations based on graph attention network and CNN-based classifier.BMC Bioinformatics. 2023 Feb 2;24(1):35. doi: 10.1186/s12859-023-05158-7. BMC Bioinformatics. 2023. PMID: 36732704 Free PMC article.
-
Prediction of microbe-drug associations based on a modified graph attention variational autoencoder and random forest.Front Microbiol. 2024 May 31;15:1394302. doi: 10.3389/fmicb.2024.1394302. eCollection 2024. Front Microbiol. 2024. PMID: 38881658 Free PMC article.
Cited by
-
Computational analysis of the gut microbiota-mediated drug metabolism.Comput Struct Biotechnol J. 2025 Mar 11;27:1472-1481. doi: 10.1016/j.csbj.2025.03.016. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40248646 Free PMC article.
References
-
- Abdoulaye O., Amadou M. L. H., Amadou O., Adakal O., Larwanou H. M., Boubou L., et al. . (2018). Epidemiological and bacteriological features of surgical site infections (ISO) in the division of surgery at the Niamey National Hospital (HNN). Pan Afr. Med. J. 31:33. doi: 10.11604/pamj.2018.31.33.15921 - DOI - PMC - PubMed
-
- Anderson T. W. (2003). An introduction to multivariate statistical analysis: United States: John Wiley&Sons.
-
- Baranwal A., Fountoulakis K., Jagannath A. (2021). Graph convolution for semisupervised classification: Improved linear separability and out-of-distribution generalization. In: International conference on machine learning (ICML). PMLR.
LinkOut - more resources
Full Text Sources

