Elucidating the structure, and composition of bacterial symbionts in the gut regions of wood-feeding termite, Coptotermes formosanus and their functional profile towards lignocellulolytic systems
- PMID: 38846576
- PMCID: PMC11155305
- DOI: 10.3389/fmicb.2024.1395568
Elucidating the structure, and composition of bacterial symbionts in the gut regions of wood-feeding termite, Coptotermes formosanus and their functional profile towards lignocellulolytic systems
Abstract
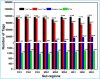
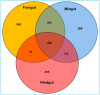
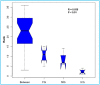
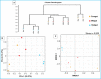
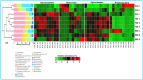
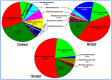
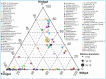
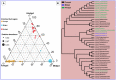
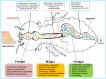
The wood-feeding termite, Coptotermes formosanus, presents an efficient lignocellulolytic system, offering a distinctive model for the exploration of host-microbial symbiosis towards lignocellulose degradation. Despite decades of investigation, understanding the diversity, community structure, and functional profiles of bacterial symbionts within specific gut regions, particularly the foregut and midgut of C. formosanus, remains largely elusive. In light of this knowledge gap, our efforts focused on elucidating the diversity, community composition and functions of symbiotic bacteria inhabiting the foregut, midgut, and hindgut of C. formosanus via metagenomics. The termite harbored a diverse community of bacterial symbionts encompassing 352 genera and 26 known phyla, exhibiting an uneven distribution across gut regions. Notably, the hindgut displayed a higher relative abundance of phyla such as Bacteroidetes (56.9%) and Spirochetes (23.3%). In contrast, the foregut and midgut were predominantly occupied by Proteobacteria (28.9%) and Firmicutes (21.2%) after Bacteroidetes. The foregut harbored unique phyla like Candidate phylum_TM6 and Armatimonadetes. At the family level, Porphyromonadaceae (28.1, 40.6, and 53.5% abundance in foregut, midgut, and hindgut, respectively) and Spirochaetaceae (foregut = 9%, midgut = 16%, hindgut = 21.6%) emerged as dominant families in the termite's gut regions. Enriched operational taxonomic units (OTUs) were most abundant in the foregut (28), followed by the hindgut (14), while the midgut exhibited enrichment of only two OTUs. Furthermore, the functional analyses revealed distinct influences of bacterial symbionts on various metabolic pathways, particularly carbohydrate and energy metabolisms of the host. Overall, these results underscore significant variations in the structure of the bacterial community among different gut regions of C. formosanus, suggesting unique functional roles of specific bacteria, thereby inspiring further investigations to resolve the crosstalk between host and microbiomes in individual gut-regions of the termite.
Keywords: bacterial diversity; gut-regions; high-throughput sequencing; microhabitats; symbiotic functions; termites.
Copyright © 2024 Dar, Xie, Jing, Qing, Ali, Pandit, Shaha and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Exploring the region-wise diversity and functions of symbiotic bacteria in the gut system of wood-feeding termite, Coptotermes formosanus, toward the degradation of cellulose, hemicellulose, and organic dyes.Insect Sci. 2022 Oct;29(5):1414-1432. doi: 10.1111/1744-7917.13012. Epub 2022 Mar 21. Insect Sci. 2022. PMID: 35134272
-
Comparison of microbial diversity and carbohydrate-active enzymes in the hindgut of two wood-feeding termites, Globitermes sulphureus (Blattaria: Termitidae) and Coptotermes formosanus (Blattaria: Rhinotermitidae).BMC Microbiol. 2024 Nov 12;24(1):470. doi: 10.1186/s12866-024-03623-8. BMC Microbiol. 2024. PMID: 39533168 Free PMC article.
-
Transcriptome analysis of the digestive system of a wood-feeding termite (Coptotermes formosanus) revealed a unique mechanism for effective biomass degradation.Biotechnol Biofuels. 2018 Feb 3;11:24. doi: 10.1186/s13068-018-1015-1. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 29434667 Free PMC article.
-
Termite symbiotic systems: efficient bio-recycling of lignocellulose.Appl Microbiol Biotechnol. 2003 Mar;61(1):1-9. doi: 10.1007/s00253-002-1189-z. Epub 2003 Jan 14. Appl Microbiol Biotechnol. 2003. PMID: 12658509 Review.
-
[Diversity and function of symbiotic microbes in the gut of lower termites].Wei Sheng Wu Xue Bao. 2006 Jun;46(3):496-9. Wei Sheng Wu Xue Bao. 2006. PMID: 16933630 Review. Chinese.
Cited by
-
Succeed to culture a novel lineage symbiotic bacterium of Mollicutes which widely found in arthropods intestine uncovers the potential double-edged sword ecological function.Front Microbiol. 2024 Oct 18;15:1458382. doi: 10.3389/fmicb.2024.1458382. eCollection 2024. Front Microbiol. 2024. PMID: 39493855 Free PMC article.
-
Phylogenetic diversity and community structure of Planctomycetota from plant biomass-rich environments.Front Microbiol. 2025 May 29;16:1579219. doi: 10.3389/fmicb.2025.1579219. eCollection 2025. Front Microbiol. 2025. PMID: 40510668 Free PMC article.
References
-
- Apprill A., McNally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microbial. Ecol. 75, 129–137. doi: 10.3354/AME01753 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

