Surveillance of vancomycin-resistant enterococci reveals shift in dominating clusters from vanA to vanB Enterococcus faecium clusters, Denmark, 2015 to 2022
- PMID: 38847117
- PMCID: PMC11158013
- DOI: 10.2807/1560-7917.ES.2024.29.23.2300633
Surveillance of vancomycin-resistant enterococci reveals shift in dominating clusters from vanA to vanB Enterococcus faecium clusters, Denmark, 2015 to 2022
Abstract
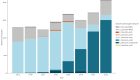
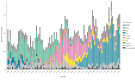
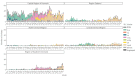
BackgroundVancomycin-resistant enterococci (VRE) are increasing in Denmark and Europe. Linezolid and vancomycin-resistant enterococci (LVRE) are of concern, as treatment options are limited. Vancomycin-variable enterococci (VVE) harbour the vanA gene complex but are phenotypically vancomycin-susceptible.AimThe aim was to describe clonal shifts for VRE and VVE in Denmark between 2015 and 2022 and to investigate genotypic linezolid resistance among the VRE and VVE.MethodsFrom 2015 to 2022, 4,090 Danish clinical VRE and VVE isolates were whole genome sequenced. We extracted vancomycin resistance genes and sequence types (STs) from the sequencing data and performed core genome multilocus sequence typing (cgMLST) analysis for Enterococcus faecium. All isolates were tested for the presence of mutations or genes encoding linezolid resistance.ResultsIn total 99% of the VRE and VVE isolates were E. faecium. From 2015 through 2019, 91.1% of the VRE and VVE were vanA E. faecium. During 2020, to the number of vanB E. faecium increased to 254 of 509 VRE and VVE isolates. Between 2015 and 2022, seven E. faecium clusters dominated: ST80-CT14 vanA, ST117-CT24 vanA, ST203-CT859 vanA, ST1421-CT1134 vanA (VVE cluster), ST80-CT1064 vanA/vanB, ST117-CT36 vanB and ST80-CT2406 vanB. We detected 35 linezolid vancomycin-resistant E. faecium and eight linezolid-resistant VVEfm.ConclusionFrom 2015 to 2022, the numbers of VRE and VVE increased. The spread of the VVE cluster ST1421-CT1134 vanA E. faecium in Denmark is a concern, especially since VVE diagnostics are challenging. The finding of LVRE, although in small numbers, ia also a concern, as treatment options are limited.
Keywords: , Enterococcus faecium; Enterococcus faecalis; LRE; LVRE; MLST; VRE; WGS; cgMLST.
Conflict of interest statement
Figures


References
-
- Pfaller MA, Cormican M, Flamm RK, Mendes RE, Jones RN. Temporal and geographic variation in antimicrobial susceptibility and resistance patterns of enterococci: results from the SENTRY antimicrobial surveillance program, 1997-2016. Open Forum Infect Dis. 2019;6(Suppl 1):S54-62. 10.1093/ofid/ofy344 - DOI - PMC - PubMed
-
- Kristich CJ, Rice LB, Arias CA. Enterococcal infection—treatment and antibiotic resistance. In: Gilmore MS, Clewell DB, Ike Y, et al, editors. Enterococci: From commensals to leading causes of drug resistant infection. Boston: Massachusetts Eye and Ear Infirmary; 2014. Available from: https://www.ncbi.nlm.nih.gov/books/NBK190420 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
