Bacterial chromatin proteins, transcription, and DNA topology: Inseparable partners in the control of gene expression
- PMID: 38847475
- PMCID: PMC11260248
- DOI: 10.1111/mmi.15283
Bacterial chromatin proteins, transcription, and DNA topology: Inseparable partners in the control of gene expression
Erratum in
-
Correction to "Bacterial Chromatin Proteins, Transcription, and DNA Topology: Inseparable Partners in the Control of Gene Expression".Mol Microbiol. 2025 Feb;123(2):176. doi: 10.1111/mmi.15324. Mol Microbiol. 2025. PMID: 39977300 No abstract available.
Abstract
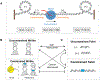
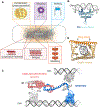
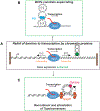
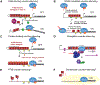
DNA in bacterial chromosomes is organized into higher-order structures by DNA-binding proteins called nucleoid-associated proteins (NAPs) or bacterial chromatin proteins (BCPs). BCPs often bind to or near DNA loci transcribed by RNA polymerase (RNAP) and can either increase or decrease gene expression. To understand the mechanisms by which BCPs alter transcription, one must consider both steric effects and the topological forces that arise when DNA deviates from its fully relaxed double-helical structure. Transcribing RNAP creates DNA negative (-) supercoils upstream and positive (+) supercoils downstream whenever RNAP and DNA are unable to rotate freely. This (-) and (+) supercoiling generates topological forces that resist forward translocation of DNA through RNAP unless the supercoiling is constrained by BCPs or relieved by topoisomerases. BCPs also may enhance topological stress and overall can either inhibit or aid transcription. Here, we review current understanding of how RNAP, BCPs, and DNA topology interplay to control gene expression.
Keywords: H‐NS; bacterial chromatin; bridging; counter‐silencing; supercoiling; topological barriers; topology; transcription.
© 2024 The Author(s). Molecular Microbiology published by John Wiley & Sons Ltd.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

