Urine scRNAseq reveals new insights into the bladder tumor immune microenvironment
- PMID: 38847806
- PMCID: PMC11157455
- DOI: 10.1084/jem.20240045
Urine scRNAseq reveals new insights into the bladder tumor immune microenvironment
Abstract
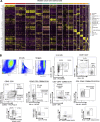
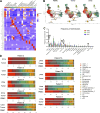
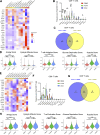
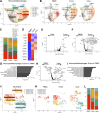
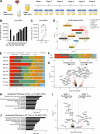
Due to bladder tumors' contact with urine, urine-derived cells (UDCs) may serve as a surrogate for monitoring the tumor microenvironment (TME) in bladder cancer (BC). However, the composition of UDCs and the extent to which they mirror the tumor remain poorly characterized. We generated the first single-cell RNA-sequencing of BC patient UDCs with matched tumor and peripheral blood mononuclear cells (PBMC). BC urine was more cellular than healthy donor (HD) urine, containing multiple immune populations including myeloid cells, CD4+ and CD8+ T cells, natural killer (NK) cells, B cells, and dendritic cells (DCs) in addition to tumor and stromal cells. Immune UDCs were transcriptionally more similar to tumor than blood. UDCs encompassed cytotoxic and activated CD4+ T cells, exhausted and tissue-resident memory CD8+ T cells, macrophages, germinal-center-like B cells, tissue-resident and adaptive NK cells, and regulatory DCs found in tumor but lacking or absent in blood. Our findings suggest BC UDCs may be surrogates for the TME and serve as therapeutic biomarkers.
© 2024 Tran et al.
Conflict of interest statement
Disclosures: R. Sebra reported being a paid consultant with GeneDx, unrelated to the submitted work. M.D. Galsky reported personal fees from Bristol Myers, Merck, Genentech, Astra Zeneca, Pfizer, EMD Serono, Seagen, Janssen, Numab, Dragonfly, Glaxo Smith Kline, Basilea, Urogen, Rappta Therapeutics, Alligator, Silverback, Fujifilm, Curis, Gilead, Bicycle, Asieris, Abbvie, Analog Devices, and Veracyte outside the submitted work; consultancy in BioMotiv, Astellas, Inctye, Dracen, Inovio, Aileron; and grants from Dendreon and Novartis. N. Bhardwaj reported “other” from DC Prime and Vaccitech and grants from Merck outside the submitted work. In addition, N. Bhardwaj receives research support from Dragonfly Therapeutics, Harbour Biomed, and Regeneron and is a consultant, advisor, or board member for Apricity, BreakBio, Carisma Therapeutics, CureVac, Genentech, Genotwin, Novartis, Primevax, Rome Therapeutics, Tempest Therapeutics, and ATP. A. Horowitz receives research funding from Astra Zeneca and has served on advisory boards for Purple Biotech and UroGen. J.P. Sfakianos reports consultancy in Pacific Edge, Natera, and Merck. No other disclosures were reported.
Figures
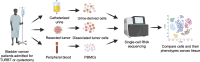








References
-
- Amara, N., Palapattu G.S., Schrage M., Gu Z., Thomas G.V., Dorey F., Said J., and Reiter R.E.. 2001. Prostate stem cell antigen is overexpressed in human transitional cell carcinoma. Cancer Res. 61:4660–4665. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

