Strategic Single-Residue Substitution in the Antimicrobial Peptide Esc(1-21) Confers Activity against Staphylococcus aureus, Including Drug-Resistant and Biofilm Phenotype
- PMID: 38848266
- PMCID: PMC11250030
- DOI: 10.1021/acsinfecdis.4c00130
Strategic Single-Residue Substitution in the Antimicrobial Peptide Esc(1-21) Confers Activity against Staphylococcus aureus, Including Drug-Resistant and Biofilm Phenotype
Abstract
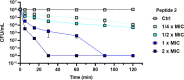
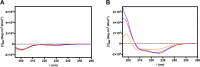
Staphylococcus aureus, a bacterium resistant to multiple drugs, is a significant cause of illness and death worldwide. Antimicrobial peptides (AMPs) provide an excellent potential strategy to cope with this threat. Recently, we characterized a derivative of the frog-skin AMP esculentin-1a, Esc(1-21) (1) that is endowed with potent activity against Gram-negative bacteria but poor efficacy against Gram-positive strains. In this study, three analogues of peptide 1 were designed by replacing Gly8 with α-aminoisobutyric acid (Aib), Pro, and dPro (2-4, respectively). The single substitution Gly8 → Aib8 in peptide 2 makes it active against the planktonic form of Gram-positive bacterial strains, especially Staphylococcus aureus, including multidrug-resistant clinical isolates, with an improved biostability without resulting in cytotoxicity to mammalian cells. Moreover, peptide 2 showed a higher antibiofilm activity than peptide 1 against both reference and clinical isolates of S. aureus. Peptide 2 was also able to induce rapid bacterial killing, suggesting a membrane-perturbing mechanism of action. Structural analysis of the most active peptide 2 evidenced that the improved biological activity of peptide 2 is the consequence of a combination of higher biostability, higher α helical content, and ability to reduce membrane fluidity and to adopt a distorted helix, bent in correspondence of Aib8. Overall, this study has shown how a strategic single amino acid substitution is sufficient to enlarge the spectrum of activity of the original peptide 1, and improve its biological properties for therapeutic purposes, thus paving the way to optimize AMPs for the development of new broad-spectrum anti-infective agents.
Keywords: Staphylococcus aureus; antimicrobial peptides; bent helical structure; biofilm; multidrug-resistant strains; α-aminoisobutyric acid.
Conflict of interest statement
The authors declare no competing financial interest.
Figures








References
-
- Romanescu M.; Oprean C.; Lombrea A.; Badescu B.; Teodor A.; Constantin G. D.; Andor M.; Folescu R.; Muntean D.; Danciu C.; Dalleur O.; Batrina S. L.; Cretu O.; Buda V. O. Current State of Knowledge Regarding WHO High Priority Pathogens-Resistance Mechanisms and Proposed Solutions through Candidates Such as Essential Oils: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 9727.10.3390/ijms24119727. - DOI - PMC - PubMed
-
- Weiner-Lastinger L. M.; Abner S.; Edwards J. R.; Kallen A. J.; Karlsson M.; Magill S. S.; Pollock D.; See I.; Soe M. M.; Walters M. S.; Dudeck M. A. Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network, 2015–2017. Infect. Control Hosp. Epidemiol. 2020, 41, 1–18. 10.1017/ice.2019.296. - DOI - PMC - PubMed
-
- Tickler I. A.; Goering R. V.; Mediavilla J. R.; Kreiswirth B. N.; Tenover F. C.; Consortium H. A. I. Continued expansion of USA300-like methicillin-resistant Staphylococcus aureus (MRSA) among hospitalized patients in the United States. Diagn. Microbiol. Infect. Dis. 2017, 88, 342–347. 10.1016/j.diagmicrobio.2017.04.016. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

