Identification of candidate chemosensory genes in the antennal transcriptome of Monolepta signata
- PMID: 38848419
- PMCID: PMC11161048
- DOI: 10.1371/journal.pone.0301177
Identification of candidate chemosensory genes in the antennal transcriptome of Monolepta signata
Abstract
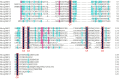
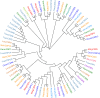
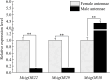
In the polyphagous insect Monolepta signata (M. signata) (Coleoptera: Chrysomelidae), antennae are important for olfactory reception used during feeding, mating, and finding a suitable oviposition site. Based on NextSeq 6000 Illumina sequencing, we assembled the antennal transcriptome of mated M. signata and described the first chemosensory gene repertoire expressed in this species. The relative expression levels of some significant chemosensory genes were conducted by quantitative real-time PCR. We identified 114 olfactory-related genes based on the antennal transcriptome database of M. signata, including 21 odorant binding proteins (OBPs), six chemosensory proteins (CSPs), 46 odorant receptors (ORs), 15 ionotropic receptors (IRs), 23 gustatory receptors (GRs) and three sensory neuron membrane proteins (SNMPs). Blastp best hit and phylogenetic analyses showed that most of the chemosensory genes had a close relationship with orthologs from other Coleoptera species. Overall, this study provides a foundation for elucidating the molecular mechanism of olfactory recognition in M. signata as well as a reference for the study of chemosensory genes in other species of Coleoptera.
Copyright: © 2024 He et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures









Similar articles
-
Antennal transcriptome analysis of olfactory genes and tissue expression profiling of odorant binding proteins in Semanotus bifasciatus (cerambycidae: coleoptera).BMC Genomics. 2022 Jun 22;23(1):461. doi: 10.1186/s12864-022-08655-w. BMC Genomics. 2022. PMID: 35733103 Free PMC article.
-
Identification of candidate chemosensory genes of Ophraella communa LeSage (Coleoptera: Chrysomelidae) based on antennal transcriptome analysis.Sci Rep. 2019 Oct 29;9(1):15551. doi: 10.1038/s41598-019-52149-x. Sci Rep. 2019. PMID: 31664149 Free PMC article.
-
Identification and characterization of chemosensory genes in the antennal transcriptome of Spodoptera exigua.Comp Biochem Physiol Part D Genomics Proteomics. 2018 Sep;27:54-65. doi: 10.1016/j.cbd.2018.05.001. Epub 2018 May 7. Comp Biochem Physiol Part D Genomics Proteomics. 2018. PMID: 29787920
-
Identification of candidate chemosensory receptors in the antennal transcriptome of the large black chafer Holotrichia parallela Motschulsky (Coleoptera: Scarabaeidae).Comp Biochem Physiol Part D Genomics Proteomics. 2018 Dec;28:63-71. doi: 10.1016/j.cbd.2018.06.005. Epub 2018 Jun 23. Comp Biochem Physiol Part D Genomics Proteomics. 2018. PMID: 29980137
-
Identification and expression analysis of candidate chemosensory receptors based on the antennal transcriptome of Lissorhoptrus oryzophilus.Comp Biochem Physiol Part D Genomics Proteomics. 2019 Jun;30:133-142. doi: 10.1016/j.cbd.2019.02.007. Epub 2019 Feb 27. Comp Biochem Physiol Part D Genomics Proteomics. 2019. PMID: 30844600
Cited by
-
Transcriptome sequencing of Antheraea pernyi antennae for identification of olfactory-related genes.BMC Genomics. 2025 May 19;26(1):499. doi: 10.1186/s12864-025-11698-4. BMC Genomics. 2025. PMID: 40383784 Free PMC article.
-
The Ultrastructure of Olfactory Sensilla Across the Antenna of Monolepta signata (Oliver).Insects. 2025 May 29;16(6):573. doi: 10.3390/insects16060573. Insects. 2025. PMID: 40559003 Free PMC article.
References
-
- Yu PY, Wang SY, Yang XK. Economic entomology of China: Volume 54, Coleptera: Chrysomeloidea (二) [M]. Beijing: Science Press, 1996. 82–169 p.
-
- Chen GH, Yin W, Li Q, Hu HY. Research progress on Monolepta hieroglyphica (Motschulsky). China Plant Prot. 2016; 36: 19–26.
-
- Tian YH, Zhang JP, Chen J, Ouyang DH, Li GW. Occurring characteristic and preventions and control strategy of Monolepta hieroglyphica (Motschulsky)—a new pest of the cotton field in Xinjiang. Anhui Agri Sci Bull. 2017; 10: 120–1+230.
-
- Du JJ, Yun L. Characteristics and control measures for the occurrence and damage of Monolepta hieroglyphica. Shaanxi J Agric Sci. 2009; 55: 202–203.
-
- Li GW, Chen XL. Studies on biological characteristics and population dynamics of Monolepta hieroglyphica in cotton in Xinjiang. China Plant Prot. 2010; 30: 8–10.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

