Strong association between genomic 3D structure and CRISPR cleavage efficiency
- PMID: 38848440
- PMCID: PMC11189236
- DOI: 10.1371/journal.pcbi.1012214
Strong association between genomic 3D structure and CRISPR cleavage efficiency
Abstract
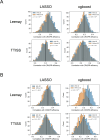
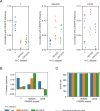
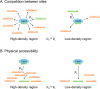
CRISPR is a gene editing technology which enables precise in-vivo genome editing; but its potential is hampered by its relatively low specificity and sensitivity. Improving CRISPR's on-target and off-target effects requires a better understanding of its mechanism and determinants. Here we demonstrate, for the first time, the chromosomal 3D spatial structure's association with CRISPR's cleavage efficiency, and its predictive capabilities. We used high-resolution Hi-C data to estimate the 3D distance between different regions in the human genome and utilized these spatial properties to generate 3D-based features, characterizing each region's density. We evaluated these features based on empirical, in-vivo CRISPR efficiency data and compared them to 425 features used in state-of-the-art models. The 3D features ranked in the top 13% of the features, and significantly improved the predictive power of LASSO and xgboost models trained with these features. The features indicated that sites with lower spatial density demonstrated higher efficiency. Understanding how CRISPR is affected by the 3D DNA structure provides insight into CRISPR's mechanism in general and improves our ability to correctly predict CRISPR's cleavage as well as design sgRNAs for therapeutic and scientific use.
Copyright: © 2024 Bergman, Tuller. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Tracking CRISPR's Footprints.Methods Mol Biol. 2019;1961:13-28. doi: 10.1007/978-1-4939-9170-9_2. Methods Mol Biol. 2019. PMID: 30912037 Review.
-
CRISPR: express delivery to any DNA address.Oral Dis. 2017 Jan;23(1):5-11. doi: 10.1111/odi.12487. Epub 2016 May 17. Oral Dis. 2017. PMID: 27040868 Review.
-
CRISPR's Path to the Clinic.CRISPR J. 2022 Feb;5(1):2-3. doi: 10.1089/crispr.2022.29143.am. CRISPR J. 2022. PMID: 35191749 No abstract available.
-
New Additions to the CRISPR Toolbox: CRISPR-CLONInG and CRISPR-CLIP for Donor Construction in Genome Editing.CRISPR J. 2020 Apr;3(2):109-122. doi: 10.1089/crispr.2019.0062. CRISPR J. 2020. PMID: 32315232 Free PMC article.
-
Codon usage and expression-based features significantly improve prediction of CRISPR efficiency.NPJ Syst Biol Appl. 2024 Sep 3;10(1):100. doi: 10.1038/s41540-024-00431-8. NPJ Syst Biol Appl. 2024. PMID: 39227603 Free PMC article.
Cited by
-
Comparison of CRISPR-Cas9, CRISPR-Cas12f1, and CRISPR-Cas3 in eradicating resistance genes KPC-2 and IMP-4.Microbiol Spectr. 2025 Jun 3;13(6):e0257224. doi: 10.1128/spectrum.02572-24. Epub 2025 Apr 28. Microbiol Spectr. 2025. PMID: 40293254 Free PMC article.
-
Characterization of the role of spatial proximity of DNA double-strand breaks in the formation of CRISPR-Cas9-induced large structural variations.Genome Res. 2025 Feb 14;35(2):231-241. doi: 10.1101/gr.278575.123. Genome Res. 2025. PMID: 39805705 Free PMC article.
References
-
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science (80-) [Internet]. 2014. Nov 28;346(6213):1258096. Available from: http://science.sciencemag.org/content/346/6213/1258096.abstract - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

