Mapping and annotating genomic loci to prioritize genes and implicate distinct polygenic adaptations for skin color
- PMID: 38849341
- PMCID: PMC11161515
- DOI: 10.1038/s41467-024-49031-4
Mapping and annotating genomic loci to prioritize genes and implicate distinct polygenic adaptations for skin color
Abstract
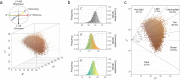
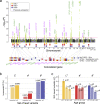
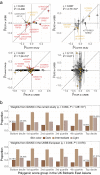
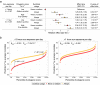
Evidence for adaptation of human skin color to regional ultraviolet radiation suggests shared and distinct genetic variants across populations. However, skin color evolution and genetics in East Asians are understudied. We quantified skin color in 48,433 East Asians using image analysis and identified associated genetic variants and potential causal genes for skin color as well as their polygenic interplay with sun exposure. This genome-wide association study (GWAS) identified 12 known and 11 previously unreported loci and SNP-based heritability was 23-24%. Potential causal genes were determined through the identification of nonsynonymous variants, colocalization with gene expression in skin tissues, and expression levels in melanocytes. Genomic loci associated with pigmentation in East Asians substantially diverged from European populations, and we detected signatures of polygenic adaptation. This large GWAS for objectively quantified skin color in an East Asian population improves understanding of the genetic architecture and polygenic adaptation of skin color and prioritizes potential causal genes.
© 2024. The Author(s).
Conflict of interest statement
Migenstory’s business is exclusively involved in providing Direct-to-Consumer (DTC) genetic testing services and generating data for research at LG H&H, without any engagement in the development of medicine or related technologies. J.G.S., S.L., H.K., K.N.G., S.W.Y., S.G.P., Y.K., and N.G.K. are employees of LG H&H. Other authors declare no other competing interests.
Figures


References
-
- Parra, E. J. Human pigmentation variation: evolution, genetic basis, and implications for public health. Am. J. Phys. Anthropol.134(Suppl 45), 85–105 (2007). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

