Additional copies of 1q negatively impact the outcome of multiple myeloma patients and induce transcriptomic deregulation in malignant plasma cells
- PMID: 38849344
- PMCID: PMC11161499
- DOI: 10.1038/s41408-024-01075-x
Additional copies of 1q negatively impact the outcome of multiple myeloma patients and induce transcriptomic deregulation in malignant plasma cells
Abstract
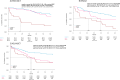
Additional copies of chromosome 1 long arm (1q) are frequently found in multiple myeloma (MM) and predict high-risk disease. Available data suggest a different outcome and biology of patients with amplification (Amp1q, ≥4 copies of 1q) vs. gain (Gain1q, 3 copies of 1q) of 1q. We evaluated the impact of Amp1q/Gain1q on the outcome of newly diagnosed MM patients enrolled in the FORTE trial (NCT02203643). Among 400 patients with available 1q data, 52 (13%) had Amp1q and 129 (32%) Gain1q. After a median follow-up of 62 months, median progression-free survival (PFS) was 21.2 months in the Amp1q group, 54.9 months in Gain1q, and not reached (NR) in Normal 1q. PFS was significantly hampered by the presence of Amp1q (HR 3.34 vs. Normal 1q, P < 0.0001; HR 1.99 vs. Gain1q, P = 0.0008). Patients with Gain1q had also a significantly shorter PFS compared with Normal 1q (HR 1.68, P = 0.0031). Concomitant poor prognostic factors or the failure to achieve MRD negativity predicted a median PFS < 12 months in Amp1q patients. Carfilzomib-lenalidomide-dexamethasone plus autologous stem cell transplantation treatment improved the adverse effect of Gain1q but not Amp1q. Transcriptomic data showed that additional 1q copies were associated with deregulation in apoptosis signaling, p38 MAPK signaling, and Myc-related genes.
© 2024. The Author(s).
Conflict of interest statement
MD has received honoraria for lectures for GlaxoSmithKline, Sanofi, and Janssen; has served on the advisory boards for GlaxoSmithKline, Sanofi, and Bristol Myers Squibb. AB has served on the advisory boards for Amgen, GlaxoSmithKline, Janssen, Pfizer. PG has served on the advisory boards for Takeda and Incyte. EZ recieved honoraria from Janssen, Bristol-Myers Squibb, Amgen, Takeda. BB has received honoraria from Amgen, Janssen, Novartis, BeiGene, Bristol Myers Squibb, GlaxoSmithKline, Jazz pharmaceuticals, Astrazeneca and Incyte; has served on the advisory boards for Amgen and Jazz pharmaceuticals. MB has received honoraria from Sanofi, Celgene, Amgen, Janssen, Novartis, Bristol Myers Squibb, and AbbVie; has served on the advisory boards for Janssen and GlaxoSmithKline; has received research funding from Sanofi, Celgene, Amgen, Janssen, Novartis, Bristol Myers Squibb, and Mundipharma. PM has received honoraria from Celgene, Janssen, Takeda, Bristol Myers Squibb, Amgen, Novartis, Gilead, Jazz, Sanofi, AbbVie, and GlaxoSmithKline; has served on the advisory boards for Celgene, Janssen, Takeda, Bristol Myers Squibb, Amgen, Jazz, Sanofi, AbbVie, and GlaxoSmithKline. FG has received honoraria from Amgen, Celgene, Janssen, Takeda, Bristol Myers Squibb, AbbVie, and GlaxoSmithKline; has served on the advisory boards for Amgen, Celgene, Janssen, Takeda, Bristol Myers Squibb, AbbVie, GlaxoSmithKline, Roche, Adaptive Biotechnologies, Oncopeptides, bluebird bio, and Pfizer. The other authors declare no competing interests.
Figures




References
-
- Perrot A, Lauwers-Cances V, Cazaubiel T, Facon T, Caillot D, Clement-Filliatre L, et al. Early versus late autologous stem cell transplant in newly diagnosed multiple myeloma: long-term follow-up analysis of the IFM 2009 trial. Blood. 2020;136:39. doi: 10.1182/blood-2020-134538. - DOI
-
- Caro J, Al Hadidi S, Usmani SZ, Yee AJ, Raje N, Davies FE. How to treat high-risk myeloma at diagnosis and relapse. In: American Society of Clinical Oncology, editor. Educational book. Alexandria (USA); vol. 2021. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical

