The role of promiscuous molecular recognition in the evolution of RNase-based self-incompatibility in plants
- PMID: 38849350
- PMCID: PMC11161657
- DOI: 10.1038/s41467-024-49163-7
The role of promiscuous molecular recognition in the evolution of RNase-based self-incompatibility in plants
Abstract
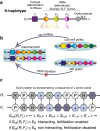
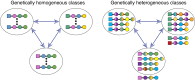
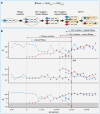
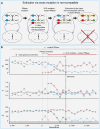
How do biological networks evolve and expand? We study these questions in the context of the plant collaborative-non-self recognition self-incompatibility system. Self-incompatibility evolved to avoid self-fertilization among hermaphroditic plants. It relies on specific molecular recognition between highly diverse proteins of two families: female and male determinants, such that the combination of genes an individual possesses determines its mating partners. Though highly polymorphic, previous models struggled to pinpoint the evolutionary trajectories by which new specificities evolved. Here, we construct a novel theoretical framework, that crucially affords interaction promiscuity and multiple distinct partners per protein, as is seen in empirical findings disregarded by previous models. We demonstrate spontaneous self-organization of the population into distinct "classes" with full between-class compatibility and a dynamic long-term balance between class emergence and decay. Our work highlights the importance of molecular recognition promiscuity to network evolvability. Promiscuity was found in additional systems suggesting that our framework could be more broadly applicable.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Insights into the Prunus-Specific S-RNase-Based Self-Incompatibility System from a Genome-Wide Analysis of the Evolutionary Radiation of S Locus-Related F-box Genes.Plant Cell Physiol. 2016 Jun;57(6):1281-94. doi: 10.1093/pcp/pcw077. Epub 2016 Apr 14. Plant Cell Physiol. 2016. PMID: 27081098
-
The proteome reveals the involvement of serine/threonine kinase in the recognition of self- incompatibility in almond.J Proteomics. 2022 Mar 30;256:104505. doi: 10.1016/j.jprot.2022.104505. Epub 2022 Feb 2. J Proteomics. 2022. PMID: 35123051
-
S-RNase evolution in self-incompatibility: Phylogenomic insights into synteny with Class I T2 RNase genes.Plant Physiol. 2025 Mar 1;197(3):kiaf072. doi: 10.1093/plphys/kiaf072. Plant Physiol. 2025. PMID: 39977114
-
Self/non-self recognition mechanisms in sexual reproduction: new insight into the self-incompatibility system shared by flowering plants and hermaphroditic animals.Biochem Biophys Res Commun. 2014 Aug 1;450(3):1142-8. doi: 10.1016/j.bbrc.2014.05.099. Epub 2014 May 27. Biochem Biophys Res Commun. 2014. PMID: 24878524 Review.
-
Self-incompatibility in Petunia: a self/nonself-recognition mechanism employing S-locus F-box proteins and S-RNase to prevent inbreeding.Wiley Interdiscip Rev Dev Biol. 2012 Mar-Apr;1(2):267-75. doi: 10.1002/wdev.10. Epub 2011 Nov 17. Wiley Interdiscip Rev Dev Biol. 2012. PMID: 23801440 Review.
References
-
- Lande R, Schemske DW. The evolution of self-fertilization and inbreeding depression in plants. I. Genetic models. Evolution. 1985;39:24–40. - PubMed
-
- de Nettancourt, D. (ed.) The genetic basis of self-incompatibility. in Incompatibility in Angiosperms, Monographs on Theoretical and Applied Genetics 28–57 (Springer, 1977). 10.1007/978-3-662-12051-4_2.
-
- Igic B, Lande R, Kohn J. Loss of self-incompatibility and its evolutionary consequences. Int. J. Plant Sci. 2008;169:93–104. doi: 10.1086/523362. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

