Revealing brain cell-stratified causality through dissecting causal variants according to their cell-type-specific effects on gene expression
- PMID: 38849352
- PMCID: PMC11161590
- DOI: 10.1038/s41467-024-49263-4
Revealing brain cell-stratified causality through dissecting causal variants according to their cell-type-specific effects on gene expression
Abstract
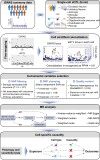
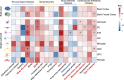
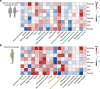
The human brain has been implicated in the pathogenesis of several complex diseases. Taking advantage of single-cell techniques, genome-wide association studies (GWAS) have taken it a step further and revealed brain cell-type-specific functions for disease loci. However, genetic causal associations inferred by Mendelian randomization (MR) studies usually include all instrumental variables from GWAS, which hampers the understanding of cell-specific causality. Here, we developed an analytical framework, Cell-Stratified MR (csMR), to investigate cell-stratified causality through colocalizing GWAS signals with single-cell eQTL from different brain cells. By applying to obesity-related traits, our results demonstrate the cell-type-specific effects of GWAS variants on gene expression, and indicate the benefits of csMR to identify cell-type-specific causal effect that is often hidden from bulk analyses. We also found csMR valuable to reveal distinct causal pathways between different obesity indicators. These findings suggest the value of our approach to prioritize target cells for extending genetic causation studies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Grants and funding
- 32170616/National Natural Science Foundation of China (National Science Foundation of China)
- 32100416/National Natural Science Foundation of China (National Science Foundation of China)
- 32070588/National Natural Science Foundation of China (National Science Foundation of China)
- 32370653/National Natural Science Foundation of China (National Science Foundation of China)
- 2021M702618/China Postdoctoral Science Foundation
LinkOut - more resources
Full Text Sources
Medical

