Protein translation rate determines neocortical neuron fate
- PMID: 38849354
- PMCID: PMC11161512
- DOI: 10.1038/s41467-024-49198-w
Protein translation rate determines neocortical neuron fate
Abstract
The mammalian neocortex comprises an enormous diversity regarding cell types, morphology, and connectivity. In this work, we discover a post-transcriptional mechanism of gene expression regulation, protein translation, as a determinant of cortical neuron identity. We find specific upregulation of protein synthesis in the progenitors of later-born neurons and show that translation rates and concomitantly protein half-lives are inherent features of cortical neuron subtypes. In a small molecule screening, we identify Ire1α as a regulator of Satb2 expression and neuronal polarity. In the developing brain, Ire1α regulates global translation rates, coordinates ribosome traffic, and the expression of eIF4A1. Furthermore, we demonstrate that the Satb2 mRNA translation requires eIF4A1 helicase activity towards its 5'-untranslated region. Altogether, we show that cortical neuron diversity is generated by mechanisms operating beyond gene transcription, with Ire1α-safeguarded proteostasis serving as an essential regulator of brain development.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
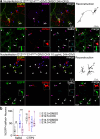
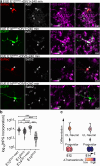
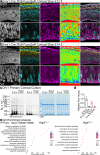





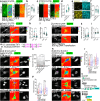
References
MeSH terms
Substances
Grants and funding
- 10.23.2.003MN/Fritz Thyssen Stiftung (Fritz Thyssen Foundation)
- 515247130/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 536563141/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 455114826/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 21-65-00017/Russian Science Foundation (RSF)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

