CellMarkerPipe: cell marker identification and evaluation pipeline in single cell transcriptomes
- PMID: 38849445
- PMCID: PMC11161599
- DOI: 10.1038/s41598-024-63492-z
CellMarkerPipe: cell marker identification and evaluation pipeline in single cell transcriptomes
Abstract
Assessing marker genes from all cell clusters can be time-consuming and lack systematic strategy. Streamlining this process through a unified computational platform that automates identification and benchmarking will greatly enhance efficiency and ensure a fair evaluation. We therefore developed a novel computational platform, cellMarkerPipe ( https://github.com/yao-laboratory/cellMarkerPipe ), for automated cell-type specific marker gene identification from scRNA-seq data, coupled with comprehensive evaluation schema. CellMarkerPipe adaptively wraps around a collection of commonly used and state-of-the-art tools, including Seurat, COSG, SC3, SCMarker, COMET, and scGeneFit. From rigorously testing across diverse samples, we ascertain SCMarker's overall reliable performance in single marker gene selection, with COSG showing commendable speed and comparable efficacy. Furthermore, we demonstrate the pivotal role of our approach in real-world medical datasets. This general and opensource pipeline stands as a significant advancement in streamlining cell marker gene identification and evaluation, fitting broad applications in the field of cellular biology and medical research.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
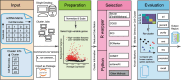
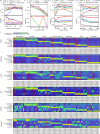
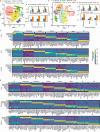
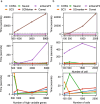
Update of
-
cellMarkerPipe: Cell Marker Identification and Evaluation Pipeline in Single Cell Transcriptomes.Res Sq [Preprint]. 2024 Jan 17:rs.3.rs-3844718. doi: 10.21203/rs.3.rs-3844718/v1. Res Sq. 2024. Update in: Sci Rep. 2024 Jun 7;14(1):13151. doi: 10.1038/s41598-024-63492-z. PMID: 38313296 Free PMC article. Updated. Preprint.
Similar articles
-
cellMarkerPipe: Cell Marker Identification and Evaluation Pipeline in Single Cell Transcriptomes.Res Sq [Preprint]. 2024 Jan 17:rs.3.rs-3844718. doi: 10.21203/rs.3.rs-3844718/v1. Res Sq. 2024. Update in: Sci Rep. 2024 Jun 7;14(1):13151. doi: 10.1038/s41598-024-63492-z. PMID: 38313296 Free PMC article. Updated. Preprint.
-
SCMarker: Ab initio marker selection for single cell transcriptome profiling.PLoS Comput Biol. 2019 Oct 28;15(10):e1007445. doi: 10.1371/journal.pcbi.1007445. eCollection 2019 Oct. PLoS Comput Biol. 2019. PMID: 31658262 Free PMC article.
-
Accurate and fast cell marker gene identification with COSG.Brief Bioinform. 2022 Mar 10;23(2):bbab579. doi: 10.1093/bib/bbab579. Brief Bioinform. 2022. PMID: 35048116
-
ASAP: a web-based platform for the analysis and interactive visualization of single-cell RNA-seq data.Bioinformatics. 2017 Oct 1;33(19):3123-3125. doi: 10.1093/bioinformatics/btx337. Bioinformatics. 2017. PMID: 28541377 Free PMC article.
-
Machine learning and statistical methods for clustering single-cell RNA-sequencing data.Brief Bioinform. 2020 Jul 15;21(4):1209-1223. doi: 10.1093/bib/bbz063. Brief Bioinform. 2020. PMID: 31243426 Review.
Cited by
-
An overview of computational methods in single-cell transcriptomic cell type annotation.Brief Bioinform. 2025 May 1;26(3):bbaf207. doi: 10.1093/bib/bbaf207. Brief Bioinform. 2025. PMID: 40347979 Free PMC article. Review.
-
scGAA: a general gated axial-attention model for accurate cell-type annotation of single-cell RNA-seq data.Sci Rep. 2024 Sep 27;14(1):22308. doi: 10.1038/s41598-024-73356-1. Sci Rep. 2024. PMID: 39333739 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

