Valine aminoacyl-tRNA synthetase promotes therapy resistance in melanoma
- PMID: 38849541
- PMCID: PMC11252002
- DOI: 10.1038/s41556-024-01439-2
Valine aminoacyl-tRNA synthetase promotes therapy resistance in melanoma
Abstract
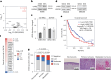
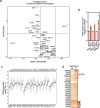
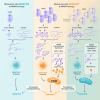
Transfer RNA dynamics contribute to cancer development through regulation of codon-specific messenger RNA translation. Specific aminoacyl-tRNA synthetases can either promote or suppress tumourigenesis. Here we show that valine aminoacyl-tRNA synthetase (VARS) is a key player in the codon-biased translation reprogramming induced by resistance to targeted (MAPK) therapy in melanoma. The proteome rewiring in patient-derived MAPK therapy-resistant melanoma is biased towards the usage of valine and coincides with the upregulation of valine cognate tRNAs and of VARS expression and activity. Strikingly, VARS knockdown re-sensitizes MAPK-therapy-resistant patient-derived melanoma in vitro and in vivo. Mechanistically, VARS regulates the messenger RNA translation of valine-enriched transcripts, among which hydroxyacyl-CoA dehydrogenase mRNA encodes for a key enzyme in fatty acid oxidation. Resistant melanoma cultures rely on fatty acid oxidation and hydroxyacyl-CoA dehydrogenase for their survival upon MAPK treatment. Together, our data demonstrate that VARS may represent an attractive therapeutic target for the treatment of therapy-resistant melanoma.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

