Extraembryonic gut endoderm cells undergo programmed cell death during development
- PMID: 38849542
- PMCID: PMC11178501
- DOI: 10.1038/s41556-024-01431-w
Extraembryonic gut endoderm cells undergo programmed cell death during development
Abstract
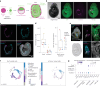
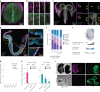
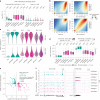
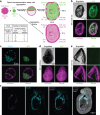
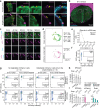
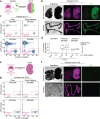
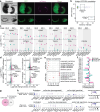
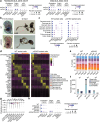
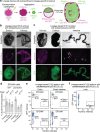
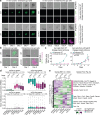
Despite a distinct developmental origin, extraembryonic cells in mice contribute to gut endoderm and converge to transcriptionally resemble their embryonic counterparts. Notably, all extraembryonic progenitors share a non-canonical epigenome, raising several pertinent questions, including whether this landscape is reset to match the embryonic regulation and if extraembryonic cells persist into later development. Here we developed a two-colour lineage-tracing strategy to track and isolate extraembryonic cells over time. We find that extraembryonic gut cells display substantial memory of their developmental origin including retention of the original DNA methylation landscape and resulting transcriptional signatures. Furthermore, we show that extraembryonic gut cells undergo programmed cell death and neighbouring embryonic cells clear their remnants via non-professional phagocytosis. By midgestation, we no longer detect extraembryonic cells in the wild-type gut, whereas they persist and differentiate further in p53-mutant embryos. Our study provides key insights into the molecular and developmental fate of extraembryonic cells inside the embryo.
© 2024. The Author(s).
Conflict of interest statement
A.M. is an inventor on a patent (US20200109456A1) related to hypermethylated CGI targets in cancer. A.M. is a co-founder and scientific advisor of Harbinger Health. The remaining authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

