DLG1 functions upstream of SDCCAG3 and IFT20 to control ciliary targeting of polycystin-2
- PMID: 38849673
- PMCID: PMC11239879
- DOI: 10.1038/s44319-024-00170-1
DLG1 functions upstream of SDCCAG3 and IFT20 to control ciliary targeting of polycystin-2
Abstract
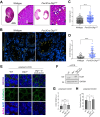
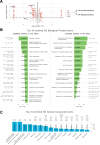
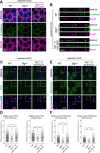
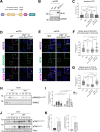
Polarized vesicular trafficking directs specific receptors and ion channels to cilia, but the underlying mechanisms are poorly understood. Here we describe a role for DLG1, a core component of the Scribble polarity complex, in regulating ciliary protein trafficking in kidney epithelial cells. Conditional knockout of Dlg1 in mouse kidney causes ciliary elongation and cystogenesis, and cell-based proximity labeling proteomics and fluorescence microscopy show alterations in the ciliary proteome upon loss of DLG1. Specifically, the retromer-associated protein SDCCAG3, IFT20, and polycystin-2 (PC2) are reduced in the cilia of DLG1-deficient cells compared to control cells. This phenotype is recapitulated in vivo and rescuable by re-expression of wild-type DLG1, but not a Congenital Anomalies of the Kidney and Urinary Tract (CAKUT)-associated DLG1 variant, p.T489R. Finally, biochemical approaches and Alpha Fold modelling suggest that SDCCAG3 and IFT20 form a complex that associates, at least indirectly, with DLG1. Our work identifies a key role for DLG1 in regulating ciliary protein composition and suggests that ciliary dysfunction of the p.T489R DLG1 variant may contribute to CAKUT.
Keywords: DLG1; IFT20; Polycystin-2; Primary Cilia; SDCCAG3.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
DLG1 functions upstream of SDCCAG3 and IFT20 to control ciliary targeting of polycystin-2.bioRxiv [Preprint]. 2024 Mar 14:2023.11.10.566524. doi: 10.1101/2023.11.10.566524. bioRxiv. 2024. Update in: EMBO Rep. 2024 Jul;25(7):3040-3063. doi: 10.1038/s44319-024-00170-1. PMID: 37987012 Free PMC article. Updated. Preprint.
References
-
- Alexa A, Rahnenfuhrer J (2023) topGO: Enrichment analysis for gene ontology, R package version 2540. p. https://bioconductor.org/packages/topGO
-
- Aslanyan MG, Doornbos C, Diwan GD, Anvarian Z, Beyer T, Junger K, van Beersum SEC, Russell RB, Ueffing M, Ludwig A, et al. A targeted multi-proteomics approach generates a blueprint of the ciliary ubiquitinome. Front Cell Dev Biol. 2023;11:1113656. doi: 10.3389/fcell.2023.1113656. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- NNF18SA0032928/Novo Nordisk Fonden (NNF)
- 20OI174/Dutch Kidney Foundation
- 3103-00177B/Danmarks Frie Forskningsfond (DFF)
- 201585/B/18/Z/Wellcome Trust (WT)
- NNF22OC0080406/Novo Nordisk Fonden (NNF)
- R01-DK108005/DK/NIDDK NIH HHS/United States
- 861329/European Union's Horizon 2020 research and innovation program Marie Sklodowska-Curie Innovative Training Networks
- R01 DK108005/DK/NIDDK NIH HHS/United States
- U54DK126114/PKD RRC
- U54 DK126114/DK/NIDDK NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- 2032-00115B/Danmarks Frie Forskningsfond (DFF)
LinkOut - more resources
Full Text Sources

