OLFM4 promotes the progression of intestinal metaplasia through activation of the MYH9/GSK3β/β-catenin pathway
- PMID: 38849840
- PMCID: PMC11157765
- DOI: 10.1186/s12943-024-02016-9
OLFM4 promotes the progression of intestinal metaplasia through activation of the MYH9/GSK3β/β-catenin pathway
Abstract
Background: Intestinal metaplasia (IM) is classified into complete intestinal metaplasia (CIM) and incomplete intestinal metaplasia (IIM). Patients diagnosed with IIM face an elevated susceptibility to the development of gastric cancer, underscoring the critical need for early screening measures. In addition to the complexities associated with diagnosis, the exact mechanisms driving the progression of gastric cancer in IIM patients remain poorly understood. OLFM4 is overexpressed in several types of tumors, including colorectal, gastric, pancreatic, and ovarian cancers, and its expression has been associated with tumor progression.
Methods: In this study, we used pathological sections from two clinical centers, biopsies of IM tissues, precancerous lesions of gastric cancer (PLGC) cell models, animal models, and organoids to explore the role of OLFM4 in IIM.
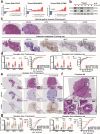
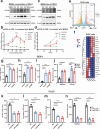
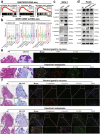
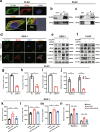
Results: Our results show that OLFM4 expression is highly increased in IIM, with superior diagnostic accuracy of IIM when compared to CDX2 and MUC2. OLFM4, along with MYH9, was overexpressed in IM organoids and PLGC animal models. Furthermore, OLFM4, in combination with Myosin heavy chain 9 (MYH9), accelerated the ubiquitination of GSK3β and resulted in increased β-catenin levels through the Wnt signaling pathway, promoting the proliferation and invasion abilities of PLGC cells.
Conclusions: OLFM4 represents a novel biomarker for IIM and could be utilized as an important auxiliary means to delimit the key population for early gastric cancer screening. Finally, our study identifies cell signaling pathways involved in the progression of IM.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Sung H, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021; 71: 209–2491. 10.3322/caac.21660. - PubMed
MeSH terms
Substances
Grants and funding
- 2021B1212040006/Guangdong Provincial Key Laboratory of Digestive Cancer Research
- 2023A1515010156/Guangdong Basic and Applied Basic Research Foundation
- JCYJ20220530144815035/the Science and Technology Planning Project of Shenzhen Municipality
- GJHZ20220913142400001/the Science and Technology Planning Project of Shenzhen Municipality
- 202104AC100001-B03/He Yulong Expert Workstation of Yunnan Province
LinkOut - more resources
Full Text Sources
Miscellaneous

