CRISPR/Cas9-induced knockout of an amino acid permease gene (AAP6) reduced Arabidopsis thaliana susceptibility to Meloidogyne incognita
- PMID: 38851681
- PMCID: PMC11162074
- DOI: 10.1186/s12870-024-05175-5
CRISPR/Cas9-induced knockout of an amino acid permease gene (AAP6) reduced Arabidopsis thaliana susceptibility to Meloidogyne incognita
Abstract
Background: Plant-parasitic root-knot nematode (Meloidogyne incognita) causes global yield loss in agri- and horticultural crops. Nematode management options rely on chemical method. However, only a handful of nematicides are commercially available. Resistance breeding efforts are not sustainable because R gene sources are limited and nematodes have developed resistance-breaking populations against the commercially available Mi-1.2 gene-expressing tomatoes. RNAi crops that manage nematode infection are yet to be commercialized because of the regulatory hurdles associated with transgenic crops. The deployment of the CRISPR/Cas9 system to improve nematode tolerance (by knocking out the susceptibility factors) in plants has emerged as a feasible alternative lately.
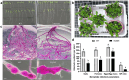
Results: In the present study, a M. incognita-responsive susceptibility (S) gene, amino acid permease (AAP6), was characterized from the model plant Arabidodpsis thaliana by generating the AtAAP6 overexpression line, followed by performing the GUS reporter assay by fusing the promoter of AtAAP6 with the β-glucuronidase (GUS) gene. Upon challenge inoculation with M. incognita, overexpression lines supported greater nematode multiplication, and AtAAP6 expression was inducible to the early stage of nematode infection. Next, using CRISPR/Cas9, AtAAP6 was selectively knocked out without incurring any growth penalty in the host plant. The 'Cas9-free' homozygous T3 line was challenge inoculated with M. incognita, and CRISPR-edited A. thaliana plants exhibited considerably reduced susceptibility to nematode infection compared to the non-edited plants. Additionally, host defense response genes were unaltered between edited and non-edited plants, implicating the direct role of AtAAP6 towards nematode susceptibility.
Conclusion: The present findings enrich the existing literature on CRISPR/Cas9 research in plant-nematode interactions, which is quite limited currently while compared with the other plant-pathogen interaction systems.
Keywords: R gene; S gene; Amino acid transporter; Gene expression; Multiplication ratio; Mutation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Functional analysis of a susceptibility gene (HIPP27) in the Arabidopsis thaliana-Meloidogyne incognita pathosystem by using a genome editing strategy.BMC Plant Biol. 2023 Aug 11;23(1):390. doi: 10.1186/s12870-023-04401-w. BMC Plant Biol. 2023. PMID: 37563544 Free PMC article.
-
The amino acid permeases AAP3 and AAP6 are involved in root-knot nematode parasitism of Arabidopsis.Mol Plant Microbe Interact. 2013 Jan;26(1):44-54. doi: 10.1094/MPMI-05-12-0123-FI. Mol Plant Microbe Interact. 2013. PMID: 23194341
-
A nematode-inducible promoter can effectively drives RNAi construct to confer Meloidogyne incognita resistance in tomato.Plant Cell Rep. 2023 Dec 20;43(1):3. doi: 10.1007/s00299-023-03114-6. Plant Cell Rep. 2023. PMID: 38117317
-
The status of the CRISPR/Cas9 research in plant-nematode interactions.Planta. 2023 Oct 24;258(6):103. doi: 10.1007/s00425-023-04259-0. Planta. 2023. PMID: 37874380 Review.
-
Silencing of disease susceptibility genes: an effective disease resistance strategy against fungal pathogens.Plant Cell Rep. 2025 May 22;44(6):127. doi: 10.1007/s00299-025-03510-0. Plant Cell Rep. 2025. PMID: 40404851 Review.
Cited by
-
Combating Root-Knot Nematodes (Meloidogyne spp.): From Molecular Mechanisms to Resistant Crops.Plants (Basel). 2025 Apr 27;14(9):1321. doi: 10.3390/plants14091321. Plants (Basel). 2025. PMID: 40364350 Free PMC article. Review.
-
Pochonia chlamydosporia synergistically supports systemic plant defense response in Phacelia tanacetifolia against Meloidogyne hapla.Front Plant Sci. 2025 Jan 16;15:1497575. doi: 10.3389/fpls.2024.1497575. eCollection 2024. Front Plant Sci. 2025. PMID: 39886679 Free PMC article.
-
Use of CRISPR Technology in Gene Editing for Tolerance to Biotic Factors in Plants: A Systematic Review.Curr Issues Mol Biol. 2024 Oct 2;46(10):11086-11123. doi: 10.3390/cimb46100659. Curr Issues Mol Biol. 2024. PMID: 39451539 Free PMC article. Review.
-
Beware of Sealing Film of Petri Dishes!-Alters the Expression of a Large Number of Genes.Int J Mol Sci. 2025 Jun 7;26(12):5484. doi: 10.3390/ijms26125484. Int J Mol Sci. 2025. PMID: 40564950 Free PMC article.
References
-
- Kumar V, Khan MR, Walia RK. Crop loss estimations due to plant-parasitic nematodes in major crops in India. Natl Acad Sci Lett. 2020;43:409–12. doi: 10.1007/s40009-020-00895-2. - DOI
-
- Phani V, Gowda MT, Dutta TK. Grafting vegetable crops to manage plant-parasitic nematodes: a review. J Pest Sci DOI. 2023 doi: 10.1007/s10340-023-01658-w. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

