Gut resistome linked to sexual preference and HIV infection
- PMID: 38851693
- PMCID: PMC11162057
- DOI: 10.1186/s12866-024-03335-z
Gut resistome linked to sexual preference and HIV infection
Abstract
Background: People living with HIV (PLWH) are at increased risk of acquisition of multidrug resistant organisms due to higher rates of predisposing factors. The gut microbiome is the main reservoir of the collection of antimicrobial resistance determinants known as the gut resistome. In PLWH, changes in gut microbiome have been linked to immune activation and HIV-1 associated complications. Specifically, gut dysbiosis defined by low microbial gene richness has been linked to low Nadir CD4 + T-cell counts. Additionally, sexual preference has been shown to strongly influence gut microbiome composition in PLWH resulting in different Prevotella or Bacteroides enriched enterotypes, in MSM (men-who-have-sex-with-men) or no-MSM, respectively. To date, little is known about gut resistome composition in PLWH due to the scarcity of studies using shotgun metagenomics. The present study aimed to detect associations between different microbiome features linked to HIV-1 infection and gut resistome composition.
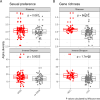
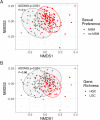
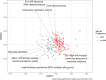
Results: Using shotgun metagenomics we characterized the gut resistome composition of 129 HIV-1 infected subjects showing different HIV clinical profiles and 27 HIV-1 negative controls from a cross-sectional observational study conducted in Barcelona, Spain. Most no-MSM showed a Bacteroides-enriched enterotype and low microbial gene richness microbiomes. We did not identify differences in resistome diversity and composition according to HIV-1 infection or immune status. However, gut resistome was more diverse in MSM group, Prevotella-enriched enterotype and gut micorbiomes with high microbial gene richness compared to no-MSM group, Bacteroides-enriched enterotype and gut microbiomes with low microbial gene richness. Additionally, gut resistome beta-diversity was different according to the defined groups and we identified a set of differentially abundant antimicrobial resistance determinants based on the established categories.
Conclusions: Our findings reveal a significant correlation between gut resistome composition and various host variables commonly associated with gut microbiome, including microbiome enterotype, microbial gene richness, and sexual preference. These host variables have been previously linked to immune activation and lower Nadir CD4 + T-Cell counts, which are prognostic factors of HIV-related comorbidities. This study provides new insights into the relationship between antibiotic resistance and clinical characteristics of PLWH.
Keywords: Antimicrobial resistance; Gut microbiome; Gut resistome; HIV infection; Shotgun metagenomics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Gut microbiota differences linked to weight gain and ART in people living with HIV are enterotype specific and minor compared to the large differences linked to sexual behavior.Front Cell Infect Microbiol. 2025 May 8;15:1568352. doi: 10.3389/fcimb.2025.1568352. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40406514 Free PMC article.
-
Gut Microbiota Linked to Sexual Preference and HIV Infection.EBioMedicine. 2016 Jan 28;5:135-46. doi: 10.1016/j.ebiom.2016.01.032. eCollection 2016 Mar. EBioMedicine. 2016. PMID: 27077120 Free PMC article.
-
Gut Microbiome Changes Associated With HIV Infection and Sexual Orientation.Front Cell Infect Microbiol. 2020 Sep 24;10:434. doi: 10.3389/fcimb.2020.00434. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33102244 Free PMC article.
-
HIV, Sexual Orientation, and Gut Microbiome Interactions.Dig Dis Sci. 2020 Mar;65(3):800-817. doi: 10.1007/s10620-020-06110-y. Dig Dis Sci. 2020. PMID: 32030625 Free PMC article. Review.
-
Helminth-Induced Human Gastrointestinal Dysbiosis: a Systematic Review and Meta-Analysis Reveals Insights into Altered Taxon Diversity and Microbial Gradient Collapse.mBio. 2021 Dec 21;12(6):e0289021. doi: 10.1128/mBio.02890-21. Epub 2021 Dec 21. mBio. 2021. PMID: 34933444 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- 847943 (MISTRAL)/European Union's Horizon 2020 Research and Innovation
- 847943 (MISTRAL)/European Union's Horizon 2020 Research and Innovation
- 847943 (MISTRAL)/European Union's Horizon 2020 Research and Innovation
- 847943 (MISTRAL)/European Union's Horizon 2020 Research and Innovation
- 847943 (MISTRAL)/European Union's Horizon 2020 Research and Innovation
LinkOut - more resources
Full Text Sources
Medical
Research Materials

