Tobacco-induced hyperglycemia promotes lung cancer progression via cancer cell-macrophage interaction through paracrine IGF2/IR/NPM1-driven PD-L1 expression
- PMID: 38851766
- PMCID: PMC11162468
- DOI: 10.1038/s41467-024-49199-9
Tobacco-induced hyperglycemia promotes lung cancer progression via cancer cell-macrophage interaction through paracrine IGF2/IR/NPM1-driven PD-L1 expression
Abstract
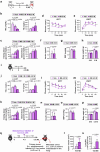
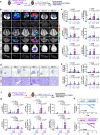
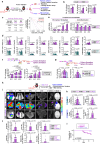
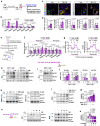
Tobacco smoking (TS) is implicated in lung cancer (LC) progression through the development of metabolic syndrome. However, direct evidence linking metabolic syndrome to TS-mediated LC progression remains to be established. Our findings demonstrate that 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and benzo[a]pyrene (NNK and BaP; NB), components of tobacco smoke, induce metabolic syndrome characteristics, particularly hyperglycemia, promoting lung cancer progression in male C57BL/6 J mice. NB enhances glucose uptake in tumor-associated macrophages by increasing the expression and surface localization of glucose transporter (GLUT) 1 and 3, thereby leading to transcriptional upregulation of insulin-like growth factor 2 (IGF2), which subsequently activates insulin receptor (IR) in LC cells in a paracrine manner, promoting its nuclear import. Nuclear IR binds to nucleophosmin (NPM1), resulting in IR/NPM1-mediated activation of the CD274 promoter and expression of programmed death ligand-1 (PD-L1). Restricting glycolysis, depleting macrophages, or blocking PD-L1 inhibits NB-mediated LC progression. Analysis of patient tissues and public databases reveals elevated levels of IGF2 and GLUT1 in tumor-associated macrophages, as well as tumoral PD-L1 and phosphorylated insulin-like growth factor 1 receptor/insulin receptor (pIGF-1R/IR) expression, suggesting potential poor prognostic biomarkers for LC patients. Our data indicate that paracrine IGF2/IR/NPM1/PD-L1 signaling, facilitated by NB-induced dysregulation of glucose levels and metabolic reprogramming of macrophages, contributes to TS-mediated LC progression.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

