Artificial intelligence-based epigenomic, transcriptomic and histologic signatures of tobacco use in oral squamous cell carcinoma
- PMID: 38851780
- PMCID: PMC11162452
- DOI: 10.1038/s41698-024-00605-x
Artificial intelligence-based epigenomic, transcriptomic and histologic signatures of tobacco use in oral squamous cell carcinoma
Abstract
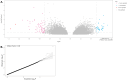
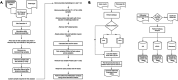
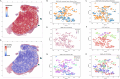
Oral squamous cell carcinoma (OSCC) biomarker studies rarely employ multi-omic biomarker strategies and pertinent clinicopathologic characteristics to predict mortality. In this study we determine for the first time a combined epigenetic, gene expression, and histology signature that differentiates between patients with different tobacco use history (heavy tobacco use with ≥10 pack years vs. no tobacco use). Using The Cancer Genome Atlas (TCGA) cohort (n = 257) and an internal cohort (n = 40), we identify 3 epigenetic markers (GPR15, GNG12, GDNF) and 13 expression markers (IGHA2, SCG5, RPL3L, NTRK1, CD96, BMP6, TFPI2, EFEMP2, RYR3, DMTN, GPD2, BAALC, and FMO3), which are dysregulated in OSCC patients who were never smokers vs. those who have a ≥ 10 pack year history. While mortality risk prediction based on smoking status and clinicopathologic covariates alone is inaccurate (c-statistic = 0.57), the combined epigenetic/expression and histologic signature has a c-statistic = 0.9409 in predicting 5-year mortality in OSCC patients.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Grants and funding
- R01 DE032501/DE/NIDCR NIH HHS/United States
- 1R01DE031395/U.S. Department of Health & Human Services | NIH | National Institute of Dental and Craniofacial Research (NIDCR)
- K23DE030250/U.S. Department of Health & Human Services | NIH | National Institute of Dental and Craniofacial Research (NIDCR)
- K23 DE030250/DE/NIDCR NIH HHS/United States
- R01DE031395/U.S. Department of Health & Human Services | NIH | National Institute of Dental and Craniofacial Research (NIDCR)
- R01 CA293995/CA/NCI NIH HHS/United States
- R56 DE030958/DE/NIDCR NIH HHS/United States
- R01 DE031395/DE/NIDCR NIH HHS/United States
- BC211095/U.S. Department of Defense (United States Department of Defense)
- R25 CA240134/CA/NCI NIH HHS/United States
- U01 DE033351/DE/NIDCR NIH HHS/United States
- L30 DE029070/DE/NIDCR NIH HHS/United States
- 1R01DE032501/U.S. Department of Health & Human Services | NIH | National Institute of Dental and Craniofacial Research (NIDCR)
- U01 CA243075/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

