Environmental microbiome diversity and stability is a barrier to antimicrobial resistance gene accumulation
- PMID: 38851788
- PMCID: PMC11162449
- DOI: 10.1038/s42003-024-06338-8
Environmental microbiome diversity and stability is a barrier to antimicrobial resistance gene accumulation
Abstract
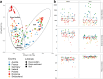
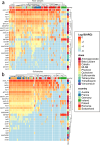
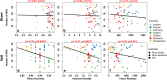
When antimicrobial resistant bacteria (ARB) and genes (ARGs) reach novel habitats, they can become part of the habitat's microbiome in the long term if they are able to overcome the habitat's biotic resilience towards immigration. This process should become more difficult with increasing biodiversity, as exploitable niches in a given habitat are reduced for immigrants when more diverse competitors are present. Consequently, microbial diversity could provide a natural barrier towards antimicrobial resistance by reducing the persistence time of immigrating ARB and ARG. To test this hypothesis, a pan-European sampling campaign was performed for structured forest soil and dynamic riverbed environments of low anthropogenic impact. In soils, higher diversity, evenness and richness were significantly negatively correlated with relative abundance of >85% of ARGs. Furthermore, the number of detected ARGs per sample were inversely correlated with diversity. However, no such effects were present in the more dynamic riverbeds. Hence, microbiome diversity can serve as a barrier towards antimicrobial resistance dissemination in stationary, structured environments, where long-term, diversity-based resilience against immigration can evolve.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
From grasslands to genes: exploring the major microbial drivers of antibiotic-resistance in microhabitats under persistent overgrazing.Microbiome. 2024 Nov 22;12(1):245. doi: 10.1186/s40168-024-01965-z. Microbiome. 2024. PMID: 39578932 Free PMC article.
-
Assessing antimicrobial resistance in pasture-based dairy farms: a 15-month surveillance study in New Zealand.Appl Environ Microbiol. 2024 Nov 20;90(11):e0139024. doi: 10.1128/aem.01390-24. Epub 2024 Oct 23. Appl Environ Microbiol. 2024. PMID: 39440981 Free PMC article.
-
Prevalence and dynamics of antimicrobial resistance in pioneer and developing Arctic soils.BMC Microbiol. 2025 Jan 27;25(1):50. doi: 10.1186/s12866-025-03745-7. BMC Microbiol. 2025. PMID: 39871128 Free PMC article.
-
Sources of Antibiotic Resistant Bacteria (ARB) and Antibiotic Resistance Genes (ARGs) in the Soil: A Review of the Spreading Mechanism and Human Health Risks.Rev Environ Contam Toxicol. 2021;256:121-153. doi: 10.1007/398_2020_60. Rev Environ Contam Toxicol. 2021. PMID: 33948742 Review.
-
Emerging soil contamination of antibiotics resistance bacteria (ARB) carrying genes (ARGs): New challenges for soil remediation and conservation.Environ Res. 2023 Feb 15;219:115132. doi: 10.1016/j.envres.2022.115132. Epub 2022 Dec 20. Environ Res. 2023. PMID: 36563979 Review.
Cited by
-
Tracing the transmission of carbapenem-resistant Enterobacterales at the patient: ward environmental nexus.Ann Clin Microbiol Antimicrob. 2024 Dec 20;23(1):108. doi: 10.1186/s12941-024-00762-8. Ann Clin Microbiol Antimicrob. 2024. PMID: 39707381 Free PMC article.
-
An eco-evolutionary perspective on antimicrobial resistance in the context of One Health.iScience. 2024 Dec 5;28(1):111534. doi: 10.1016/j.isci.2024.111534. eCollection 2025 Jan 17. iScience. 2024. PMID: 39801834 Free PMC article. Review.
-
Contrary effects of increasing temperatures on the spread of antimicrobial resistance in river biofilms.mSphere. 2024 Feb 28;9(2):e0057323. doi: 10.1128/msphere.00573-23. Epub 2024 Feb 7. mSphere. 2024. PMID: 38323843 Free PMC article.
-
Resistome in the indoor dust samples from workplaces and households: a pilot study.Front Cell Infect Microbiol. 2024 Dec 3;14:1484100. doi: 10.3389/fcimb.2024.1484100. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39691696 Free PMC article.
-
Species-resolved profiling of antibiotic resistance genes in complex metagenomes through long-read overlapping with Argo.Nat Commun. 2025 Feb 18;16(1):1744. doi: 10.1038/s41467-025-57088-y. Nat Commun. 2025. PMID: 39966439 Free PMC article.
References
-
- WHO. Antimicrobial Resistance Global Report on Surveillance. (2014).
-
- WHO. Global action plan on antimicrobial resistance. Microbe Mag. 10, 354–355 (2015).
Publication types
MeSH terms
Substances
Grants and funding
- I 4374/FWF_/Austrian Science Fund FWF/Austria
- 202004910327/China Scholarship Council (CSC)
- 01LC1904A/Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research)
- I 4374-B/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- 186531/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- 460816351/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 01DO2200/Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research)
- ANR-19-EBI3-0005-04/Agence Nationale de la Recherche (French National Research Agency)
- 2019-NC-MS-9/Environmental Protection Agency (EPA)
LinkOut - more resources
Full Text Sources
Medical

