Investigating the potential of 6-substituted 3-formyl chromone derivatives as anti-diabetic agents using in silico methods
- PMID: 38851807
- PMCID: PMC11162442
- DOI: 10.1038/s41598-024-63237-y
Investigating the potential of 6-substituted 3-formyl chromone derivatives as anti-diabetic agents using in silico methods
Abstract
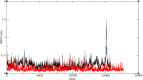
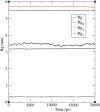
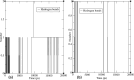
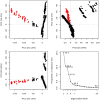
In exploring nature's potential in addressing diabetes-related conditions, this study investigates the therapeutic capabilities of 3-formyl chromone derivatives. Utilizing in silico methodologies, we focus on 6-substituted 3-formyl chromone derivatives (1-16) to assess their therapeutic potential in treating diabetes. The research examined the formyl group at the chromone's C-3 position. ADMET, biological activities, were conducted along with B3LYP calculations using 3 different basis sets. The analogues were analyzed based on their parent structure obtained from PubChem. The HOMO-LUMO gap confirmed the bioactive nature of the derivatives, NBO analysis was performed to understand the charge transfer. PASS prediction revealed that 3-formyl chromone derivatives are potent aldehyde oxidase inhibitors, insulin inhibitors, HIF1A expression inhibitors, and histidine kinase. Molecular docking studies indicated that the compounds had a strong binding affinity with proteins, including CAD, BHK, IDE, HIF-α, p53, COX, and Mpro of SARS-CoV2. 6-isopropyl-3-formyl chromone (4) displayed the highest affinity for IDE, with a binding energy of - 8.5 kcal mol-1. This result outperformed the affinity of the reference standard dapagliflozin (- 7.9 kcal mol-1) as well as two other compounds that target human IDE, namely vitexin (- 8.3 kcal mol-1) and myricetin (- 8.4 kcal mol-1). MD simulations were revealed RMSD value between 0.2 and 0.5 nm, indicating the strength of the protein-ligand complex at the active site.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

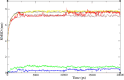

Similar articles
-
Synthesis, Molecular Docking, Molecular Dynamics Studies, and Biological Evaluation of 4H-Chromone-1,2,3,4-tetrahydropyrimidine-5-carboxylate Derivatives as Potential Antileukemic Agents.J Chem Inf Model. 2017 Jun 26;57(6):1246-1257. doi: 10.1021/acs.jcim.6b00138. Epub 2017 May 25. J Chem Inf Model. 2017. PMID: 28524659
-
Chromone-embedded peptidomimetics and furopyrimidines as highly potent SARS-CoV-2 infection inhibitors: docking and MD simulation study.BMC Res Notes. 2023 Sep 21;16(1):224. doi: 10.1186/s13104-023-06508-7. BMC Res Notes. 2023. PMID: 37735703 Free PMC article.
-
Exploring the effectiveness of flavone derivatives for treating liver diseases: Utilizing DFT, molecular docking, and molecular dynamics techniques.MethodsX. 2023 Dec 29;12:102537. doi: 10.1016/j.mex.2023.102537. eCollection 2024 Jun. MethodsX. 2023. PMID: 38299040 Free PMC article.
-
Investigation of the New Inhibitors by Sulfadiazine and Modified Derivatives of α-D-glucopyranoside for White Spot Syndrome Virus Disease of Shrimp by In Silico: Quantum Calculations, Molecular Docking, ADMET and Molecular Dynamics Study.Molecules. 2022 Jun 8;27(12):3694. doi: 10.3390/molecules27123694. Molecules. 2022. PMID: 35744817 Free PMC article.
-
Identification of a potential SARS-CoV2 inhibitor via molecular dynamics simulations and amino acid decomposition analysis.J Biomol Struct Dyn. 2021 Oct;39(17):6633-6648. doi: 10.1080/07391102.2020.1797536. Epub 2020 Jul 24. J Biomol Struct Dyn. 2021. PMID: 32705953 Free PMC article.
Cited by
-
Exploring the Therapeutic Potential of Petiveria alliacea L. Phytochemicals: A Computational Study on Inhibiting SARS-CoV-2's Main Protease (Mpro).Molecules. 2024 May 27;29(11):2524. doi: 10.3390/molecules29112524. Molecules. 2024. PMID: 38893400 Free PMC article.
References
-
- Kazeem M, Bankole H, Ogunrinola O, Wusu A, Kappo A. Functional foods with dipeptidyl peptidase-4 inhibitory potential and management of type 2 diabetes: A review. Food Front. 2021;2:153–162. doi: 10.1002/fft2.71. - DOI
-
- Yuan J, et al. Single-junction organic solar cell with over 15% efficiency using fused-ring acceptor with electron-deficient core. Joule. 2019;3:1140–1151. doi: 10.1016/j.joule.2019.01.004. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

