Ratiometric nonfluorescent CRISPR assay utilizing Cas12a-induced plasmid supercoil relaxation
- PMID: 38851849
- PMCID: PMC11162422
- DOI: 10.1038/s42004-024-01214-2
Ratiometric nonfluorescent CRISPR assay utilizing Cas12a-induced plasmid supercoil relaxation
Abstract
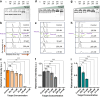
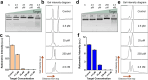
Most CRISPR-based biosensors rely on labeled reporter molecules and expensive equipment for signal readout. A recent approach quantifies analyte concentration by sizing λ DNA reporters via gel electrophoresis, providing a simple solution for label-free detection. Here, we report an alternative strategy for label-free CRISPR-Cas12a, which relies on Cas12a trans-nicking induced supercoil relaxation of dsDNA plasmid reporters to generate a robust and ratiometric readout. The ratiometric CRISPR (rCRISPR) measures the relative percentage of supercoiled plasmid DNA to the relaxed circular DNA by gel electrophoresis for more accurate target concentration quantification. This simple method is two orders of magnitude more sensitive than the typical fluorescent reporter. This self-referenced strategy solves the potential application limitations of previously demonstrated DNA sizing-based CRISPR-Dx without compromising the sensitivity. Finally, we demonstrated the applicability of rCRISPR for detecting various model DNA targets such as HPV 16 and real AAV samples, highlighting its feasibility for point-of-care CRISPR-Dx applications.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
A Sensitive and Nonoptical CRISPR Detection Mechanism by Sizing Double-Stranded λ DNA Reporter.Angew Chem Int Ed Engl. 2022 Dec 12;61(50):e202213920. doi: 10.1002/anie.202213920. Epub 2022 Nov 10. Angew Chem Int Ed Engl. 2022. PMID: 36239984 Free PMC article.
-
Ratiometric fluorescent probe: a sensitive and reliable reporter for the CRISPR/Cas12a-based biosensing platform.Analyst. 2022 May 30;147(11):2567-2574. doi: 10.1039/d2an00613h. Analyst. 2022. PMID: 35575140
-
Unidirectional trans-cleaving behavior of CRISPR-Cas12a unlocks for an ultrasensitive assay using hybrid DNA reporters containing a 3' toehold.Nucleic Acids Res. 2023 Oct 13;51(18):9894-9904. doi: 10.1093/nar/gkad715. Nucleic Acids Res. 2023. PMID: 37650631 Free PMC article.
-
Sensitive and Portable Signal Readout Strategies Boost Point-of-Care CRISPR/Cas12a Biosensors.ACS Sens. 2023 Nov 24;8(11):3988-4007. doi: 10.1021/acssensors.3c01338. Epub 2023 Oct 23. ACS Sens. 2023. PMID: 37870387 Review.
-
Towards application of CRISPR-Cas12a in the design of modern viral DNA detection tools (Review).J Nanobiotechnology. 2022 Jan 21;20(1):41. doi: 10.1186/s12951-022-01246-7. J Nanobiotechnology. 2022. PMID: 35062978 Free PMC article. Review.
Cited by
-
Attomolar Detection of HIV-1 with Label-Free RCA-rCRISPR on Smartphone.bioRxiv [Preprint]. 2025 Jun 17:2025.06.15.659809. doi: 10.1101/2025.06.15.659809. bioRxiv. 2025. PMID: 40667362 Free PMC article. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources

