Immune profiling of age and adjuvant-specific activation of human blood mononuclear cells in vitro
- PMID: 38851856
- PMCID: PMC11162429
- DOI: 10.1038/s42003-024-06390-4
Immune profiling of age and adjuvant-specific activation of human blood mononuclear cells in vitro
Abstract
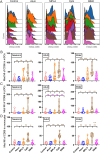
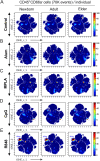
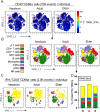
Vaccination reduces morbidity and mortality due to infections, but efficacy may be limited due to distinct immunogenicity at the extremes of age. This raises the possibility of employing adjuvants to enhance immunogenicity and protection. Early IFNγ production is a hallmark of effective vaccine immunogenicity in adults serving as a biomarker that may predict effective adjuvanticity. We utilized mass cytometry (CyTOF) to dissect the source of adjuvant-induced cytokine production in human blood mononuclear cells (BMCs) from newborns (~39-week-gestation), adults (~18-63 years old) and elders (>65 years of age) after stimulation with pattern recognition receptors agonist (PRRa) adjuvants. Dimensionality reduction analysis of CyTOF data mapped the BMC compartment, elucidated age-specific immune responses and profiled PRR-mediated activation of monocytes and DCs upon adjuvant stimulation. Furthermore, we demonstrated PRRa adjuvants mediated innate IFNγ induction and mapped NK cells as the key source of TLR7/8 agonist (TLR7/8a) specific innate IFNγ responses. Hierarchical clustering analysis revealed age and TLR7/8a-specific accumulation of innate IFNγ producing γδ T cells. Our study demonstrates the application of mass cytometry and cutting-edge computational approaches to characterize immune responses across immunologically distinct age groups and may inform identification of the bespoke adjuvantation systems tailored to enhance immunity in distinct vulnerable populations.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing financial interest(s): D.S., O.L. and D.J.D. are named inventors on patents relating to small molecule adjuvants assigned to Boston Children’s Hospital. O.L.’s laboratory has received sponsored research support from
Figures




Similar articles
-
Adjuvant Effect of Bacille Calmette-Guérin on Hepatitis B Vaccine Immunogenicity in the Preterm and Term Newborn.Front Immunol. 2018 Jan 24;9:29. doi: 10.3389/fimmu.2018.00029. eCollection 2018. Front Immunol. 2018. PMID: 29416539 Free PMC article.
-
Adjuvant-induced Human Monocyte Secretome Profiles Reveal Adjuvant- and Age-specific Protein Signatures.Mol Cell Proteomics. 2016 Jun;15(6):1877-94. doi: 10.1074/mcp.M115.055541. Epub 2016 Mar 1. Mol Cell Proteomics. 2016. PMID: 26933193 Free PMC article.
-
Adjuvant-Associated Peripheral Blood mRNA Profiles and Kinetics Induced by the Adjuvanted Recombinant Protein Candidate Tuberculosis Vaccine M72/AS01 in Bacillus Calmette-Guérin-Vaccinated Adults.Front Immunol. 2018 Mar 26;9:564. doi: 10.3389/fimmu.2018.00564. eCollection 2018. Front Immunol. 2018. PMID: 29632533 Free PMC article. Clinical Trial.
-
Toward precision adjuvants: optimizing science and safety.Curr Opin Pediatr. 2020 Feb;32(1):125-138. doi: 10.1097/MOP.0000000000000868. Curr Opin Pediatr. 2020. PMID: 31904601 Free PMC article. Review.
-
Adjuvant system AS01: from mode of action to effective vaccines.Expert Rev Vaccines. 2024 Jan-Dec;23(1):715-729. doi: 10.1080/14760584.2024.2382725. Epub 2024 Aug 5. Expert Rev Vaccines. 2024. PMID: 39042099 Review.
Cited by
-
Immune profiling reveals umbilical cord blood mononuclear cells from South India display an IL-8 dominant, CXCL-10 deficient polyfunctional monocyte response to pathogen-associated molecular patterns that is distinct from adult blood cells.Clin Exp Immunol. 2024 Aug 9;217(3):263-278. doi: 10.1093/cei/uxae034. Clin Exp Immunol. 2024. PMID: 38695079 Free PMC article.
-
The recent advances in vaccine adjuvants.Front Immunol. 2025 May 13;16:1557415. doi: 10.3389/fimmu.2025.1557415. eCollection 2025. Front Immunol. 2025. PMID: 40433383 Free PMC article. Review.
-
Immune Evasion in Cancer Metastasis: An Unappreciated Role of Monocytes.Cancers (Basel). 2025 May 12;17(10):1638. doi: 10.3390/cancers17101638. Cancers (Basel). 2025. PMID: 40427136 Free PMC article. Review.
-
Nonhuman primate models of pediatric viral diseases.Front Cell Infect Microbiol. 2024 Dec 3;14:1493885. doi: 10.3389/fcimb.2024.1493885. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39691699 Free PMC article. Review.
References
-
- Donald, K. & Finlay, B. B. Early-life interactions between the microbiota and immune system: impact on immune system development and atopic disease. Nat. Rev. Immunol.23, 735–748 (2023). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 75N93019C00044/AI/NIAID NIH HHS/United States
- 272201800047C-P00003-9999)/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- 272201800047C/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources

