Identifying the molecular basis of Laminin N-terminal domain Ca2+ binding using a hybrid approach
- PMID: 38851889
- PMCID: PMC11365105
- DOI: 10.1016/j.bpj.2024.06.005
Identifying the molecular basis of Laminin N-terminal domain Ca2+ binding using a hybrid approach
Abstract
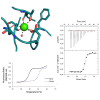
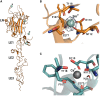
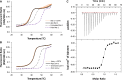
Ca2+ is a highly abundant ion involved in numerous biological processes, particularly in multicellular eukaryotic organisms where it exerts many of these functions through interactions with Ca2+ binding proteins. The laminin N-terminal (LN) domain is found in members of the laminin and netrin protein families where it plays a critical role in the function of these proteins. The LN domain of laminins and netrins is a Ca2+ binding domain and in many cases requires Ca2+ to perform its biological function. Here, we conduct a detailed examination of the molecular basis of the LN domain Ca2+ interaction combining structural, computational, bioinformatics, and biophysical techniques. By combining computational and bioinformatic techniques with x-ray crystallography we explore the molecular basis of the LN domain Ca2+ interaction and identify a conserved sequence present in Ca2+ binding LN domains. These findings enable a sequence-based prediction of LN domain Ca2+ binding ability. We use thermal shift assays and isothermal titration calorimetry to explore the biophysical properties of the LN domain Ca2+ interaction. We show that the netrin-1 LN domain exhibits a high affinity and specificity for Ca2+, which structurally stabilizes the LN domain. This study elucidates the molecular foundation of the LN domain Ca2+ binding interaction and provides a detailed functional characterization of this essential interaction, advancing our understanding of protein-Ca2+ dynamics within the context of the LN domain.
Copyright © 2024 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declared no competing interests.
Figures

References
-
- Carafoli E., Santella L., et al. Brini M. Generation, control, and processing of cellular calcium signals. Crit. Rev. Biochem. Mol. Biol. 2001;36:107–260. - PubMed
-
- Clapham D.E. Calcium Signaling. Cell. 2007;131:1047–1058. - PubMed
-
- Brown E.M., MacLeod R.J. Extracellular calcium sensing and extracellular calcium signaling. Physiol. Rev. 2001;81:239–297. - PubMed
-
- Handschuh G., Candidus S., et al. Becker K.-F. Tumour-associated E-cadherin mutations alter cellular morphology, decrease cellular adhesion and increase cellular motility. Oncogene. 1999;18:4301–4312. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

