Cellular-resolution gene expression mapping reveals organization in the head ganglia of the gastropod, Berghia stephanieae
- PMID: 38852042
- PMCID: PMC11198006
- DOI: 10.1002/cne.25628
Cellular-resolution gene expression mapping reveals organization in the head ganglia of the gastropod, Berghia stephanieae
Abstract
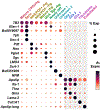
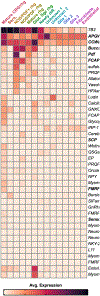
Gastropod molluscs such as Aplysia, Lymnaea, and Tritonia have been important for determining fundamental rules of motor control, learning, and memory because of their large, individually identifiable neurons. Yet only a small number of gastropod neurons have known molecular markers, limiting the ability to establish brain-wide structure-function relations. Here we combine high-throughput, single-cell RNA sequencing with in situ hybridization chain reaction in the nudibranch Berghia stephanieae to identify and visualize the expression of markers for cell types. Broad neuronal classes were characterized by genes associated with neurotransmitters, like acetylcholine, glutamate, serotonin, and GABA, as well as neuropeptides. These classes were subdivided by other genes including transcriptional regulators and unannotated genes. Marker genes expressed by neurons and glia formed discrete, previously unrecognized regions within and between ganglia. This study provides the foundation for understanding the fundamental cellular organization of gastropod nervous systems.
Keywords: Identified neurons; glial cells; neuroanatomy; neurochemistry; transcription factors.
© 2024 The Author(s). The Journal of Comparative Neurology published by Wiley Periodicals LLC.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures
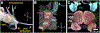
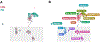

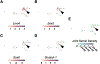




Similar articles
-
Egg-laying hormone expression in identified neurons across developmental stages and reproductive states of the nudibranch Berghia stephanieae.Horm Behav. 2024 Aug;164:105578. doi: 10.1016/j.yhbeh.2024.105578. Epub 2024 Jun 25. Horm Behav. 2024. PMID: 38925074 Free PMC article.
-
Expression patterns and behavioral effects of conopressin and APGWamide in the nudibranch Berghia stephanieae.Peptides. 2024 Sep;179:171253. doi: 10.1016/j.peptides.2024.171253. Epub 2024 May 29. Peptides. 2024. PMID: 38821120
-
Comparative mapping of GABA-immunoreactive neurons in the central nervous systems of nudibranch molluscs.J Comp Neurol. 2014 Mar;522(4):794-810. doi: 10.1002/cne.23446. J Comp Neurol. 2014. PMID: 24638845
-
The importance of identified neurons in gastropod molluscs to neuroscience.Curr Opin Neurobiol. 2019 Jun;56:1-7. doi: 10.1016/j.conb.2018.10.009. Epub 2018 Nov 1. Curr Opin Neurobiol. 2019. PMID: 30390485 Review.
-
The pharmacology of molluscan neurons.Prog Neurobiol. 1984;23(1-2):79-150. doi: 10.1016/0301-0082(84)90013-3. Prog Neurobiol. 1984. PMID: 6151715 Review.
Cited by
-
Functional mapping of the molluscan brain guided by synchrotron X-ray tomography.Proc Natl Acad Sci U S A. 2025 Mar 4;122(9):e2422706122. doi: 10.1073/pnas.2422706122. Epub 2025 Feb 27. Proc Natl Acad Sci U S A. 2025. PMID: 40014565 Free PMC article.
-
Circuits and the single neuron.J Neurophysiol. 2024 Sep 1;132(3):765-766. doi: 10.1152/jn.00312.2024. Epub 2024 Jul 31. J Neurophysiol. 2024. PMID: 39081212 Free PMC article. No abstract available.
-
The legacies of A. O. Dennis Willows and Peter A. Getting: neuroscience research using Tritonia.J Neurophysiol. 2025 Jan 1;133(1):34-45. doi: 10.1152/jn.00318.2024. Epub 2024 Nov 29. J Neurophysiol. 2025. PMID: 39611858 Review.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ. 1990. “Basic Local Alignment Search Tool.” J. Mol. Biol 215 (3): 403–10. - PubMed
-
- Arey LB (1918). THE MULTIPLE SENSORY ACTIVITIES OF THE SO-CALLED RHINOPHORE OF NUDIBRANCHS. American Journal of Physiology-Legacy Content, 46(5), 526–532. 10.1152/ajplegacy.1918.46.5.526 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

