Comprehensive functional characterization of complement factor I rare variant genotypes identified in the SCOPE geographic atrophy cohort
- PMID: 38852887
- PMCID: PMC11277764
- DOI: 10.1016/j.jbc.2024.107452
Comprehensive functional characterization of complement factor I rare variant genotypes identified in the SCOPE geographic atrophy cohort
Abstract
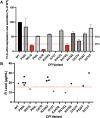
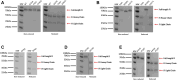
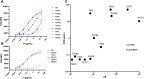
Rare variants (RVs) in the gene encoding the regulatory enzyme complement factor I (CFI; FI) that reduce protein function or levels increase age-related macular degeneration risk. A total of 3357 subjects underwent screening in the SCOPE natural history study for geographic atrophy secondary to age-related macular degeneration, including CFI sequencing and serum FI measurement. Eleven CFI RV genotypes that were challenging to categorize as type I (low serum level) or type II (normal serum level, reduced enzymatic function) were characterized in the context of pure FI protein in C3b and C4b fluid phase cleavage assays and a novel bead-based functional assay (BBFA) of C3b cleavage. Four variants predicted or previously characterized as benign were analyzed by BBFA for comparison. In all, three variants (W51S, C67R, and I370T) resulted in low expression. Furthermore, four variants (P64L, R339Q, G527V, and P528T) were identified as being highly deleterious with IC50s for C3b breakdown >1 log increased versus the WT protein, while two variants (K476E and R474Q) were ∼1 log reduced in function. Meanwhile, six variants (P50A, T203I, K441R, E548Q, P553S, and S570T) had IC50s similar to WT. Odds ratios and BBFA IC50s were positively correlated (r = 0.76, p < 0.01), while odds ratios versus combined annotation dependent depletion (CADD) scores were not (r = 0.43, p = 0.16). Overall, 15 CFI RVs were functionally characterized which may aid future patient stratification for complement-targeted therapies. Pure protein in vitro analysis remains the gold standard for determining the functional consequence of CFI RVs.
Keywords: C3 glomerulopathy(C3G); age-related macular degeneration (AMD); atypical hemolytic uremic syndrome (aHUS); complement; complement factor I; complement system; enzyme mutation; innate immunity; retinal degeneration.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest D. K.: Gyroscope Therapeutics (consultancy, equity, grant income), Novartis (consultancy), Alexion Pharmaceuticals (consultancy), Apellis (consultancy); Sarepta (consultancy), Chemocentryx (Consultancy), Sobi (consultancy), Samsung (consultancy), Purespring (consultancy), Roche (consultancy); C. L. H: Q32 Bio (consultancy), Gyroscope Therapeutics (Consultancy), Novartis (employee), Ra Pharmaceuticals (Grant income), Biocryst (consultancy); K. J. M: Qualasept (consultancy), Freeline Therapeutics (consultancy), Catalyst Biosciences (grant income, consultancy), Idorsia Pharmaceuticals (grant income), Gemini Therapeutics (grant income, consultancy), Alexion Pharmaceuticals (grant income, consultancy); T. M. H.: Gyroscope Therapeutics, Novartis (employee); S. J. S.: Gyroscope Therapeutics, Novartis (employee); E. G.: Gyroscope Therapeutics, Novartis (employee); A. D.: Gyroscope Therapeutics, Novartis, Beacon Therapeutics (employee); A. V. J.: Gyroscope Therapeutics, Novartis (employee); A. L.: Gyroscope Therapeutics (consultancy, equity), Roche (consultancy), Apellis (consultancy), Novartis (consultancy).
Figures



References
-
- Wong W.L., Su X., Li X., Cheung C.M.G., Klein R., Cheng C.Y., et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob. Health. 2014;2:e106–e116. - PubMed
-
- Wang Y., Zhong Y., Zhang L., Wu Q., Tham Y., Rim T.H., et al. Global incidence, progression, and risk factors of age-related macular degeneration and projection of disease Statistics in 30 Years: a modeling study. Gerontology. 2022;68:721–735. - PubMed
-
- Fleckenstein M., Keenan T.D.L., Guymer R.H., Chakravarthy U., Schmitz-Valckenberg S., Klaver C.C., et al. Age-related macular degeneration. Nat. Rev. Dis. Primers. 2021;7:31. - PubMed
-
- Maguire M.G., Martin D.F., Ying G.S., Jaffe G.J., Daniel E., Grunwald J.E., et al. Five-year outcomes with anti–Vascular Endothelial Growth factor treatment of Neovascular age-related macular degeneration: the comparison of age-related macular degeneration treatments trials. Ophthalmology. 2016;123:1751–1761. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

