RET overexpression leads to increased brain metastatic competency in luminal breast cancer
- PMID: 38852945
- PMCID: PMC11461165
- DOI: 10.1093/jnci/djae091
RET overexpression leads to increased brain metastatic competency in luminal breast cancer
Abstract
Background: Breast cancer brain metastasis is a rising occurrence, necessitating a better understanding of the mechanisms involved for effective management. Breast cancer brain metastases diverge notably from the primary tumor, with gains in kinase and concomitant losses of steroid signaling observed. In this study, we explored the role of the kinase receptor RET in promoting breast cancer brain metastases and provide a rationale for targeting this receptor.
Methods: RET expression was characterized in a cohort of patients with primary and brain metastatic tumors. RET functionality was assessed using pharmacological inhibition and gene silencing in patient-derived brain metastatic tumor explants and in vivo models, organoid models, and brain organotypic cultures. RNA sequencing was used to uncover novel brain metastatic relevant RET mechanisms of action.
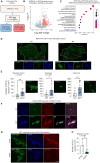
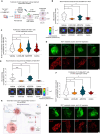
Results: A statistically significant enrichment of RET in brain metastases was observed in estrogen receptor-positive breast cancer, where it played a role in promoting cancer cell adhesion, survival, and outgrowth in the brain. In vivo, RET overexpression enhanced brain metastatic competency in patient-derived models. At a mechanistic level, RET overexpression was found to enhance the activation of gene programs involved in cell adhesion, requiring EGFR cooperation to deliver a pro-brain metastatic phenotype.
Conclusion: Our results illustrate, for the first time, the role of RET in regulating colonization and outgrowth of breast cancer brain metastasis and provide data to support the use of RET inhibitors in the management strategy for patients with breast cancer brain metastases.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Leone JP, Lin NU.. Systemic therapy of central nervous system metastases of breast cancer. Curr Oncol Rep. 2019;21(7):59. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

