Deep learning models for predicting the survival of patients with hepatocellular carcinoma based on a surveillance, epidemiology, and end results (SEER) database analysis
- PMID: 38853169
- PMCID: PMC11163004
- DOI: 10.1038/s41598-024-63531-9
Deep learning models for predicting the survival of patients with hepatocellular carcinoma based on a surveillance, epidemiology, and end results (SEER) database analysis
Abstract
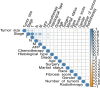
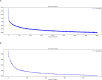
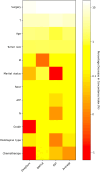
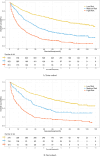
Hepatocellular carcinoma (HCC) is a common malignancy with poor survival and requires long-term follow-up. Hence, we collected information on patients with Primary Hepatocellular Carcinoma in the United States from the Surveillance, Epidemiology, and EndResults (SEER) database. We used this information to establish a deep learning with a multilayer neural network (the NMTLR model) for predicting the survival rate of patients with Primary Hepatocellular Carcinoma. HCC patients pathologically diagnosed between January 2011 and December 2015 in the SEER (Surveillance, Epidemiology, and End Results) database of the National Cancer Institute of the United States were selected as study subjects. We utilized two deep learning-based algorithms (DeepSurv and Neural Multi-Task Logistic Regression [NMTLR]) and a machine learning-based algorithm (Random Survival Forest [RSF]) for model training. A multivariable Cox Proportional Hazards (CoxPH) model was also constructed for comparison. The dataset was randomly divided into a training set and a test set in a 7:3 ratio. The training dataset underwent hyperparameter tuning through 1000 iterations of random search and fivefold cross-validation. Model performance was assessed using the concordance index (C-index), Brier score, and Integrated Brier Score (IBS). The accuracy of predicting 1-year, 3-year, and 5-year survival rates was evaluated using Receiver Operating Characteristic (ROC) curves, calibration plots, and Area Under the Curve (AUC). The primary outcomes were the 1-year, 3-year, and 5-year overall survival rates. Models were developed using DeepSurv, NMTLR, RSF, and Cox Proportional Hazards regression. Model differentiation was evaluated using the C-index, calibration with concordance plots, and risk stratification capability with the log-rank test. The study included 2197 HCC patients, randomly divided into a training cohort (70%, n = 1537) and a testing cohort (30%, n = 660). Clinical characteristics between the two cohorts showed no significant statistical difference (p > 0.05). The deep learning models outperformed both RSF and CoxPH models, with C-indices of 0.735 (NMTLR) and 0.731 (DeepSurv) in the test dataset. The NMTLR model demonstrated enhanced accuracy and well-calibrated survival estimates, achieving an Area Under the Curve (AUC) of 0.824 for 1-year survival predictions, 0.813 for 3-year, and 0.803 for 5-year survival rates. This model's superior calibration and discriminative ability enhance its utility for clinical prognostication in Primary Hepatocellular Carcinoma. We deployed the NMTLR model as a web application for clinical practice. The NMTLR model have potential advantages over traditional linear models in prognostic assessment and treatment recommendations. This novel analytical approach may provide reliable information on individual survival and treatment recommendations for patients with primary liver cancer.
Keywords: Deep learning; Machine learning; Predictive model; Primary liver cancer; SEER.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Deep learning models for predicting the survival of patients with chondrosarcoma based on a surveillance, epidemiology, and end results analysis.Front Oncol. 2022 Aug 22;12:967758. doi: 10.3389/fonc.2022.967758. eCollection 2022. Front Oncol. 2022. PMID: 36072795 Free PMC article.
-
Development and validation of a deep learning-based survival prediction model for pediatric glioma patients: A retrospective study using the SEER database and Chinese data.Comput Biol Med. 2024 Nov;182:109185. doi: 10.1016/j.compbiomed.2024.109185. Epub 2024 Sep 27. Comput Biol Med. 2024. PMID: 39341114
-
Machine learning-based individualized survival prediction model for prognosis in osteosarcoma: Data from the SEER database.Medicine (Baltimore). 2024 Sep 27;103(39):e39582. doi: 10.1097/MD.0000000000039582. Medicine (Baltimore). 2024. PMID: 39331900 Free PMC article.
-
The power of deep learning in simplifying feature selection for hepatocellular carcinoma: a review.BMC Med Inform Decis Mak. 2024 Oct 4;24(1):287. doi: 10.1186/s12911-024-02682-1. BMC Med Inform Decis Mak. 2024. PMID: 39367397 Free PMC article. Review.
-
Application of machine learning in predicting survival outcomes involving real-world data: a scoping review.BMC Med Res Methodol. 2023 Nov 13;23(1):268. doi: 10.1186/s12874-023-02078-1. BMC Med Res Methodol. 2023. PMID: 37957593 Free PMC article.
Cited by
-
Concordance-based Predictive Uncertainty (CPU)-Index: Proof-of-concept with application towards improved specificity of lung cancers on low dose screening CT.Artif Intell Med. 2025 Feb;160:103055. doi: 10.1016/j.artmed.2024.103055. Epub 2024 Dec 16. Artif Intell Med. 2025. PMID: 39721356
-
Deep neural network provides personalized treatment recommendations for de novo metastatic breast cancer patients.J Cancer. 2024 Oct 28;15(20):6668-6685. doi: 10.7150/jca.101293. eCollection 2024. J Cancer. 2024. PMID: 39668839 Free PMC article.
-
Predicting the prognosis of epithelial ovarian cancer patients based on deep learning models.Front Oncol. 2025 Jul 25;15:1592746. doi: 10.3389/fonc.2025.1592746. eCollection 2025. Front Oncol. 2025. PMID: 40786516 Free PMC article.
References
-
- Sung, H., Ferlay, J., Siegel, R. L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. (2021). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

