Cell surface CD55 traffics to the nucleus leading to cisplatin resistance and stemness by inducing PRC2 and H3K27 trimethylation on chromatin in ovarian cancer
- PMID: 38853277
- PMCID: PMC11163727
- DOI: 10.1186/s12943-024-02028-5
Cell surface CD55 traffics to the nucleus leading to cisplatin resistance and stemness by inducing PRC2 and H3K27 trimethylation on chromatin in ovarian cancer
Abstract
Background: Platinum resistance is the primary cause of poor survival in ovarian cancer (OC) patients. Targeted therapies and biomarkers of chemoresistance are critical for the treatment of OC patients. Our previous studies identified cell surface CD55, a member of the complement regulatory proteins, drives chemoresistance and maintenance of cancer stem cells (CSCs). CSCs are implicated in tumor recurrence and metastasis in multiple cancers.
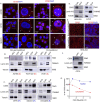
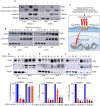
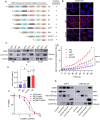
Methods: Protein localization assays including immunofluorescence and subcellular fractionation were used to identify CD55 at the cell surface and nucleus of cancer cells. Protein half-life determinations were used to compare cell surface and nuclear CD55 stability. CD55 deletion mutants were generated and introduced into cancer cells to identify the nuclear trafficking code, cisplatin sensitivity, and stem cell frequency that were assayed using in vitro and in vivo models. Detection of CD55 binding proteins was analyzed by immunoprecipitation followed by mass spectrometry. Target pathways activated by CD55 were identified by RNA sequencing.
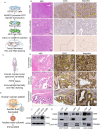
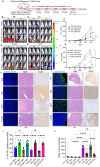
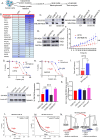
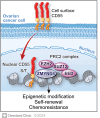
Results: CD55 localizes to the nucleus of a subset of OC specimens, ascites from chemoresistant patients, and enriched in chemoresistant OC cells. We determined that nuclear CD55 is glycosylated and derived from the cell surface pool of CD55. Nuclear localization is driven by a trafficking code containing the serine/threonine (S/T) domain of CD55. Nuclear CD55 is necessary for cisplatin resistance, stemness, and cell proliferation in OC cells. CD55 S/T domain is necessary for nuclear entry and inducing chemoresistance to cisplatin in both in vitro and in vivo models. Deletion of the CD55 S/T domain is sufficient to sensitize chemoresistant OC cells to cisplatin. In the nucleus, CD55 binds and attenuates the epigenetic regulator and tumor suppressor ZMYND8 with a parallel increase in H3K27 trimethylation and members of the Polycomb Repressive Complex 2.
Conclusions: For the first time, we show CD55 localizes to the nucleus in OC and promotes CSC and chemoresistance. Our studies identify a therapeutic mechanism for treating platinum resistant ovarian cancer by blocking CD55 nuclear entry.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

