A comprehensive Thai pharmacogenomics database (TPGxD-1): Phenotype prediction and variants identification in 942 whole-genome sequencing data
- PMID: 38853370
- PMCID: PMC11163017
- DOI: 10.1111/cts.13830
A comprehensive Thai pharmacogenomics database (TPGxD-1): Phenotype prediction and variants identification in 942 whole-genome sequencing data
Abstract
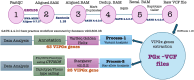
Computational methods analyze genomic data to identify genetic variants linked to drug responses, thereby guiding personalized medicine. This study analyzed 942 whole-genome sequences from the Electricity Generating Authority of Thailand (EGAT) cohort to establish a population-specific pharmacogenomic database (TPGxD-1) in the Thai population. Sentieon (version 201808.08) implemented the GATK best workflow practice for variant calling. We then annotated variant call format (VCF) files using Golden Helix VarSeq 2.5.0 and employed Stargazer v2.0.2 for star allele analysis. The analysis of 63 very important pharmacogenes (VIPGx) reveals 85,566 variants, including 13,532 novel discoveries. Notably, we identified 464 known PGx variants and 275 clinically relevant novel variants. The phenotypic prediction of 15 VIPGx demonstrated a varied metabolic profile for the Thai population. Genes like CYP2C9 (9%), CYP3A5 (45.2%), CYP2B6 (9.4%), NUDT15 (15%), CYP2D6 (47%) and CYP2C19 (43%) showed a high number of intermediate metabolizers; CYP3A5 (41%), and CYP2C19 (9.9%) showed more poor metabolizers. CYP1A2 (52.7%) and CYP2B6 (7.6%) were found to have a higher number of ultra-metabolizers. The functional prediction of the remaining 10 VIPGx genes reveals a high frequency of decreased functional alleles in SULT1A1 (12%), NAT2 (84%), and G6PD (12%). SLCO1B1 reports 20% poor functional alleles, while PTGIS (42%), SLCO1B1 (4%), and TPMT (5.96%) showed increased functional alleles. This study discovered new variants and alleles in the 63 VIPGx genes among the Thai population, offering insights into advancing clinical pharmacogenomics (PGx). However, further validation is needed using other computational and genotyping methods.
© 2024 The Author(s). Clinical and Translational Science published by Wiley Periodicals LLC on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
The authors declared no competing interests for this work.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous