Targeting IGF2 to reprogram the tumor microenvironment for enhanced viro-immunotherapy
- PMID: 38853689
- PMCID: PMC11376453
- DOI: 10.1093/neuonc/noae105
Targeting IGF2 to reprogram the tumor microenvironment for enhanced viro-immunotherapy
Abstract
Background: The FDA approval of oncolytic herpes simplex-1 virus (oHSV) therapy underscores its therapeutic promise and safety as a cancer immunotherapy. Despite this promise, the current efficacy of oHSV is significantly limited to a small subset of patients largely due to the resistance in tumor and tumor microenvironment (TME).
Methods: RNA sequencing (RNA-Seq) was used to identify molecular targets of oHSV resistance. Intracranial human and murine glioma or breast cancer brain metastasis (BCBM) tumor-bearing mouse models were employed to elucidate the mechanism underlying oHSV therapy-induced resistance.
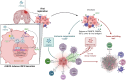
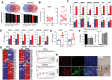
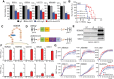
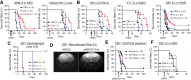
Results: Transcriptome analysis identified IGF2 as one of the top-secreted proteins following oHSV treatment. Moreover, IGF2 expression was significantly upregulated in 10 out of 14 recurrent GBM patients after treatment with oHSV, rQNestin34.5v.2 (71.4%; P = .0020) (ClinicalTrials.gov, NCT03152318). Depletion of IGF2 substantially enhanced oHSV-mediated tumor cell killing in vitro and improved survival of mice bearing BCBM tumors in vivo. To mitigate the oHSV-induced IGF2 in the TME, we constructed a novel oHSV, oHSV-D11mt, secreting a modified IGF2R domain 11 (IGF2RD11mt) that acts as IGF2 decoy receptor. Selective blocking of IGF2 by IGF2RD11mt significantly increased cytotoxicity, reduced oHSV-induced neutrophils/PMN-MDSCs infiltration, and reduced secretion of immune suppressive/proangiogenic cytokines, while increased CD8 + cytotoxic T lymphocytes (CTLs) infiltration, leading to enhanced survival in GBM or BCBM tumor-bearing mice.
Conclusions: This is the first study reporting that oHSV-induced secreted IGF2 exerts a critical role in resistance to oHSV therapy, which can be overcome by oHSV-D11mt as a promising therapeutic advance for enhanced viro-immunotherapy.
Keywords: Oncolytic herpes simplex virus-1 (oHSV); glioblastoma (GBM); insulin-like growth factor 2 (IGF2); insulin-like growth factor-1 receptor (IGF1R); tumor microenvironment (TME).
© The Author(s) 2024. Published by Oxford University Press on behalf of the Society for Neuro-Oncology.
Conflict of interest statement
The authors have no potential conflicts of interest to disclose.
Figures


References
-
- Arefian E, Bamdad T, Soleimanjahi H, et al. Evaluation of gamma-interferon kinetics in HSV-1 infected mice in different days post infection (in vivo) and post re-stimulation (in vitro). Comp Immunol Microbiol Infect Dis. 2007;30(1):1–9. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

