Integrating Clinical Phenotype With Multiomics Analyses of Human Cardiac Tissue Unveils Divergent Metabolic Remodeling in Genotype-Positive and Genotype-Negative Patients With Hypertrophic Cardiomyopathy
- PMID: 38853772
- PMCID: PMC11188634
- DOI: 10.1161/CIRCGEN.123.004369
Integrating Clinical Phenotype With Multiomics Analyses of Human Cardiac Tissue Unveils Divergent Metabolic Remodeling in Genotype-Positive and Genotype-Negative Patients With Hypertrophic Cardiomyopathy
Abstract
Background: Hypertrophic cardiomyopathy (HCM) is caused by sarcomere gene mutations (genotype-positive HCM) in ≈50% of patients and occurs in the absence of mutations (genotype-negative HCM) in the other half of patients. We explored how alterations in the metabolomic and lipidomic landscape are involved in cardiac remodeling in both patient groups.
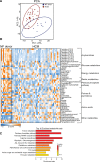
Methods: We performed proteomics, metabolomics, and lipidomics on myectomy samples (genotype-positive N=19; genotype-negative N=22; and genotype unknown N=6) from clinically well-phenotyped patients with HCM and on cardiac tissue samples from sex- and age-matched and body mass index-matched nonfailing donors (N=20). These data sets were integrated to comprehensively map changes in lipid-handling and energy metabolism pathways. By linking metabolomic and lipidomic data to variability in clinical data, we explored patient group-specific associations between cardiac and metabolic remodeling.
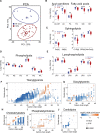
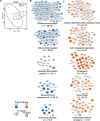
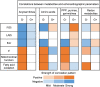
Results: HCM myectomy samples exhibited (1) increased glucose and glycogen metabolism, (2) downregulation of fatty acid oxidation, and (3) reduced ceramide formation and lipid storage. In genotype-negative patients, septal hypertrophy and diastolic dysfunction correlated with lowering of acylcarnitines, redox metabolites, amino acids, pentose phosphate pathway intermediates, purines, and pyrimidines. In contrast, redox metabolites, amino acids, pentose phosphate pathway intermediates, purines, and pyrimidines were positively associated with septal hypertrophy and diastolic impairment in genotype-positive patients.
Conclusions: We provide novel insights into both general and genotype-specific metabolic changes in HCM. Distinct metabolic alterations underlie cardiac disease progression in genotype-negative and genotype-positive patients with HCM.
Keywords: cardiomyopathies; genotype; hypertrophy; lipidomics; metabolism; metabolomics.
Conflict of interest statement
Figures




References
-
- Semsarian C, Ingles J, Maron MS, Maron BJ. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol. 2015;65:1249–1254. doi: 10.1016/j.jacc.2015.01.019 - PubMed
-
- Ommen SR, Mital S, Burke MA, Day SM, Deswal A, Elliott P, Evanovich LL, Hung J, Joglar JA, Kantor P, et al. . 2020 AHA/ACC guideline for the diagnosis and treatment of patients with hypertrophic cardiomyopathy: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J Am Coll Cardiol. 2020;76:e159–e240. doi: 10.1016/j.jacc.2020.08.045 - PubMed
-
- Maron BJ. Clinical course and management of hypertrophic cardiomyopathy. N Engl J Med. 2018;379:1977. doi: 10.1056/NEJMc1812159 - PubMed
-
- Neubauer S, Kolm P, Ho CY, Kwong RY, Desai MY, Dolman SF, Appelbaum E, Desvigne-Nickens P, DiMarco JP, Friedrich MG, et al. ; HCMR Investigators. Distinct subgroups in hypertrophic cardiomyopathy in the NHLBI HCM registry. J Am Coll Cardiol. 2019;74:2333–2345. doi: 10.1016/j.jacc.2019.08.1057 - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

