This is a preprint.
Structural dynamics of the intrinsically disordered linker region of cardiac troponin T
- PMID: 38853835
- PMCID: PMC11160775
- DOI: 10.1101/2024.05.30.596451
Structural dynamics of the intrinsically disordered linker region of cardiac troponin T
Abstract
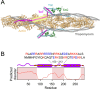
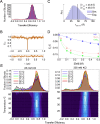
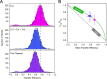
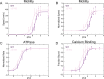
The cardiac troponin complex, composed of troponins I, T, and C, plays a central role in regulating the calcium-dependent interactions between myosin and the thin filament. Mutations in troponin can cause cardiomyopathies; however, it is still a major challenge to connect how changes in sequence affect troponin's function. Recent high-resolution structures of the thin filament revealed critical insights into the structure-function relationship of troponin, but there remain large, unresolved segments of troponin, including the troponin-T linker region that is a hotspot for cardiomyopathy mutations. This linker region is predicted to be intrinsically disordered, with behaviors that are not well described by traditional structural approaches; however, this proposal has not been experimentally verified. Here, we used a combination of single-molecule Förster resonance energy transfer (FRET), molecular dynamics simulations, and functional reconstitution assays to investigate the troponin-T linker region. We show that in the context of both isolated troponin and the fully regulated troponin complex, the linker behaves as a dynamic, intrinsically disordered region. This region undergoes polyampholyte expansion in the presence of high salt and distinct conformational changes during the assembly of the troponin complex. We also examine the ΔE160 hypertrophic cardiomyopathy mutation in the linker and demonstrate that it does not affect the conformational dynamics of the linker, rather it allosterically affects interactions with other troponin complex subunits, leading to increased molecular contractility. Taken together, our data clearly demonstrate the importance of disorder within the troponin-T linker and provide new insights into the molecular mechanisms driving the pathogenesis of cardiomyopathies.
Keywords: Major Classification: Biological Sciences; Minor Classification: Biophysics and Computational Biology; sarcomere; single molecule; thin filament.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT All experiments were conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest. M.J.G. acknowledges research support from Edgewise Therapeutics unrelated to this project.
Figures
Similar articles
-
FRET-based analysis of the cardiac troponin T linker region reveals the structural basis of the hypertrophic cardiomyopathy-causing Δ160E mutation.J Biol Chem. 2019 Oct 4;294(40):14634-14647. doi: 10.1074/jbc.RA118.005098. Epub 2019 Aug 6. J Biol Chem. 2019. PMID: 31387947 Free PMC article.
-
Troponins, intrinsic disorder, and cardiomyopathy.Biol Chem. 2016 Aug 1;397(8):731-51. doi: 10.1515/hsz-2015-0303. Biol Chem. 2016. PMID: 27074551
-
Computational Studies of Cardiac and Skeletal Troponin.Front Mol Biosci. 2019 Aug 9;6:68. doi: 10.3389/fmolb.2019.00068. eCollection 2019. Front Mol Biosci. 2019. PMID: 31448287 Free PMC article. Review.
-
Structure and Dynamics of the Flexible Cardiac Troponin T Linker Domain in a Fully Reconstituted Thin Filament.Biochemistry. 2022 Jul 5;61(13):1229-1242. doi: 10.1021/acs.biochem.2c00091. Epub 2022 Jun 13. Biochemistry. 2022. PMID: 35696530 Free PMC article.
-
Mouse Models of Cardiomyopathies Caused by Mutations in Troponin C.Int J Mol Sci. 2023 Aug 2;24(15):12349. doi: 10.3390/ijms241512349. Int J Mol Sci. 2023. PMID: 37569724 Free PMC article. Review.
References
-
- Willott R. H., et al., Mutations in Troponin that cause HCM, DCM AND RCM: what can we learn about thin filament function? J. Mol. Cell. Cardiol. 48, 882–892 (2010). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
