This is a preprint.
Prokrustean Graph: A substring index for rapid k-mer size analysis
- PMID: 38853857
- PMCID: PMC11160577
- DOI: 10.1101/2023.11.21.568151
Prokrustean Graph: A substring index for rapid k-mer size analysis
Abstract
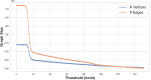
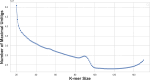
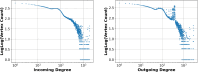
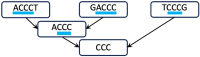
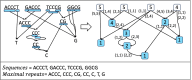
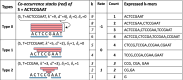
The widespread adoption of k-mers in bioinformatics has led to efficient methods utilizing genomic sequences in a variety of biological tasks. However, understanding the influence of k-mer sizes within these methods remains a persistent challenge, as the outputs of complex bioinformatics pipelines obscure this influence with various noisy factors. The choice of k-mer size is often arbitrary, with justification frequently omitted in the literature and method tutorials. Furthermore, recent methods employing multiple k-mer sizes encounter significant computational challenges. Nevertheless, most methods are built on well-defined objects related to k-mers, such as de Bruijn graphs, Jaccard similarity, Bray-Curtis dissimilarity, and k-mer spectra. The role of k-mer sizes within these objects is more intuitive and can be described by numerous quantities and metrics. Therefore, exploring these objects across k-mer sizes opens opportunities for robust analyses and new applications. However, the evolution of k-mer objects with respect to k-mer sizes is surprisingly elusive. We introduce a novel substring index, the Prokrustean graph, that elucidates the transformation of k-mer sets across k-mer sizes. Our framework built upon this index rapidly computes k-mer-based quantities for all k-mer sizes, with computational complexity independent of the size range and dependent only on maximal repeats. For example, counting maximal simple paths in de Bruijn graphs for is achieved in seconds using our index on a gigabase-scale dataset. We present a variety of such experiments relevant to pangenomics and metagenomics. The Prokrustean graph is space-efficiently constructed from the Burrows-Wheeler Transform. Through this construction, it becomes evident that other modern substring indices inherently face difficulties in exploring k-mer objects across sizes, which motivated our data structure. Our implementation is available at: https://github.com/KoslickiLab/prokrustean.
Figures


Similar articles
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Systemic Inflammatory Response Syndrome.2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31613449 Free Books & Documents.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Bankevich A., Bzikadze A. V., Kolmogorov M., Antipov D., and Pevzner P. A.. Multiplex de bruijn graphs enable genome assembly from long, high-fidelity reads. Nature biotechnology, 40(7):1075–1081, 2022. - PubMed
-
- Belazzougui D.. Linear time construction of compressed text indices in compact space. In Proceedings of the forty-sixth Annual ACM Symposium on Theory of Computing, pages 148–193, 2014.
-
- Belazzougui D. and Cunial F.. Fully-functional bidirectional burrows-wheeler indexes and infinite-order de bruijn graphs. In 30th Annual Symposium on Combinatorial Pattern Matching (CPM 2019). Schloss Dagstuhl-Leibniz-Zentrum fuer Informatik, 2019.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
