This is a preprint.
A glial circadian gene expression atlas reveals cell type and disease-specific reprogramming in response to amyloid pathology or aging
- PMID: 38853870
- PMCID: PMC11160685
- DOI: 10.1101/2024.05.28.596297
A glial circadian gene expression atlas reveals cell type and disease-specific reprogramming in response to amyloid pathology or aging
Update in
-
A glial circadian gene expression atlas reveals cell-type and disease-specific reprogramming in response to amyloid pathology or aging.Nat Neurosci. 2025 Nov;28(11):2366-2379. doi: 10.1038/s41593-025-02067-1. Epub 2025 Oct 23. Nat Neurosci. 2025. PMID: 41131361 Free PMC article.
Abstract
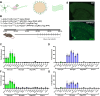
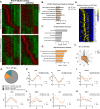
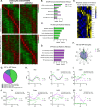
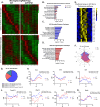
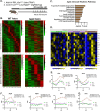
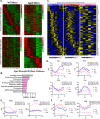
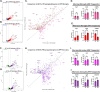
While circadian rhythm disruption may promote neurodegenerative disease, how aging and neurodegenerative pathology impact circadian gene expression patterns in different brain cell types is unknown. Here, we used translating ribosome affinity purification methods to define the circadian translatomes of astrocytes, microglia, and bulk cerebral cortex, in healthy mouse brain and in the settings of amyloid-beta plaque pathology or aging. Our data reveal that glial circadian translatomes are highly cell type-specific and exhibit profound, context-dependent reprogramming of rhythmic transcripts in response to amyloid pathology or aging. Transcripts involved in glial activation, immunometabolism, and proteostasis, as well as nearly half of all Alzheimer Disease (AD)-associated risk genes, displayed circadian oscillations, many of which were altered by pathology. Amyloid-related differential gene expression was also dependent on time of day. Thus, circadian rhythms in gene expression are cell- and context dependent and provide important insights into glial gene regulation in health, AD, and aging.
Keywords: Alzheimer’s Disease; aging; astrocyte; circadian; glia; microglia; transcriptome.
Figures
References
-
- Zhang R., Lahens N.F., Ballance H.I., Hughes M.E., and Hogenesch J.B. (2014). A circadian gene expression atlas in mammals: Implications for biology and medicine. Proceedings of the National Academy of Sciences of the United States of America 111, 16219–16224. 10.1073/pnas.1408886111. - DOI - PMC - PubMed
-
- Panda S., Antoch M.P., Miller B.H., Su A.I., Schook A.B., Straume M., Schultz P.G., Kay S.A., Takahashi J.S., and Hogenesch J.B. (2002). Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320. - PubMed
-
- Debski K.J., Ceglia N., Ghestem A., Ivanov A.I., Brancati G.E., Bröer S., Bot A.M., Müller J.A., Schoch S., Becker A., et al. (2020). The circadian dynamics of the hippocampal transcriptome and proteome is altered in experimental temporal lobe epilepsy. Sci Adv 6. 10.1126/sciadv.aat5979. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
