This is a preprint.
The Iroquois (Iro/Irx) homeobox genes are conserved Hox targets involved in motor neuron development
- PMID: 38853975
- PMCID: PMC11160718
- DOI: 10.1101/2024.05.30.596714
The Iroquois (Iro/Irx) homeobox genes are conserved Hox targets involved in motor neuron development
Update in
-
The Iroquois (Iro/Irx) homeobox genes are conserved Hox targets involved in motor neuron development.iScience. 2025 Mar 12;28(4):112210. doi: 10.1016/j.isci.2025.112210. eCollection 2025 Apr 18. iScience. 2025. PMID: 40230530 Free PMC article.
Abstract
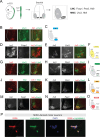
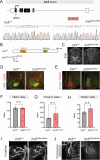
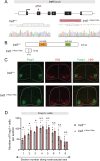
The Iroquois (Iro/Irx) homeobox genes encode transcription factors with fundamental roles in animal development. Despite their link to various congenital conditions in humans, our understanding of Iro/Irx gene expression, function, and regulation remains incomplete. Here, we conducted a systematic expression analysis of all six mouse Irx genes in the embryonic spinal cord. We found five Irx genes (Irx1, Irx2, Irx3, Irx5, and Irx6) to be confined mostly to ventral spinal domains, offering new molecular markers for specific groups of post-mitotic motor neurons (MNs). Further, we engineered Irx2, Irx5, and Irx6 mouse mutants and uncovered essential but distinct roles for Irx2 and Irx6 in MN development. Last, we found that the highly conserved regulators of MN development across species, the HOX proteins, directly control Irx gene expression both in mouse and C. elegans MNs, critically expanding the repertoire of HOX target genes in the developing nervous system. Altogether, our study provides important insights into Iro/Irx expression and function in the developing spinal cord, and uncovers an ancient gene regulatory relationship between HOX and Iro/Irx genes.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures




Similar articles
-
The Iroquois (Iro/Irx) homeobox genes are conserved Hox targets involved in motor neuron development.iScience. 2025 Mar 12;28(4):112210. doi: 10.1016/j.isci.2025.112210. eCollection 2025 Apr 18. iScience. 2025. PMID: 40230530 Free PMC article.
-
Expression of two novel mouse Iroquois homeobox genes during neurogenesis.Mech Dev. 2000 Mar 1;91(1-2):317-21. doi: 10.1016/s0925-4773(99)00263-4. Mech Dev. 2000. PMID: 10704856
-
The Xenopus Irx genes are essential for neural patterning and define the border between prethalamus and thalamus through mutual antagonism with the anterior repressors Fezf and Arx.Dev Biol. 2009 May 15;329(2):258-68. doi: 10.1016/j.ydbio.2009.02.028. Epub 2009 Mar 4. Dev Biol. 2009. PMID: 19268445
-
The Role of IRX Homeobox Genes in Hematopoietic Progenitors and Leukemia.Genes (Basel). 2023 Jan 23;14(2):297. doi: 10.3390/genes14020297. Genes (Basel). 2023. PMID: 36833225 Free PMC article. Review.
-
Iroquois genes: genomic organization and function in vertebrate neural development.Curr Opin Genet Dev. 2002 Aug;12(4):403-8. doi: 10.1016/s0959-437x(02)00317-9. Curr Opin Genet Dev. 2002. PMID: 12100884 Review.
References
-
- Cavodeassi F., Modolell J. & Gomez-Skarmeta J. L. The Iroquois family of genes: from body building to neural patterning. Development 128, 2847–2855 (2001). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
