This is a preprint.
The Latent Aging of Cells
- PMID: 38854054
- PMCID: PMC11160607
- DOI: 10.1101/2024.05.28.596284
The Latent Aging of Cells
Abstract
As epigenetic clocks have evolved from powerful estimators of chronological aging to predictors of mortality and disease risk, it begs the question of what role DNA methylation plays in the aging process. We hypothesize that while it has the potential to serve as an informative biomarker, DNA methylation could also be a key to understanding the biology entangled between aging, (de)differentiation, and epigenetic reprogramming. Here we use an unsupervised approach to analyze time associated DNA methylation from both in vivo and in vitro samples to measure an underlying signal that ties these phenomena together. We identify a methylation pattern shared across all three, as well as a signal that tracks aging in tissues but appears refractory to reprogramming, suggesting that aging and reprogramming may not be fully mirrored processes.
Conflict of interest statement
Declarations All authors are employees of Altos Labs.
Figures


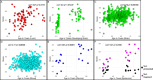




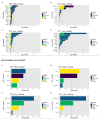
References
-
- Ahuja N, Q Li A Msohan L, Baylin S B, and Issa J P. 1998. “Aging and DNA Methylation in Colorectal Mucosa and Cancer.” Cancer Research 58 (23): 5489–94. - PubMed
-
- Bernhart Stephan H., Kretzmer Helene, Holdt Lesca M., Frank Jühling Ole Ammerpohl, Bergmann Anke K., Northoff Bernd H., et al. 2016. “Changes of Bivalent Chromatin Coincide with Increased Expression of Developmental Genes in Cancer.” Scientific Reports 6 (1): 37393. 10.1038/srep37393. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
