This is a preprint.
Distinct mechanisms of non-autonomous UPRER mediated by GABAergic, glutamatergic, and octopaminergic neurons
- PMID: 38854121
- PMCID: PMC11160609
- DOI: 10.1101/2024.05.27.595950
Distinct mechanisms of non-autonomous UPRER mediated by GABAergic, glutamatergic, and octopaminergic neurons
Abstract
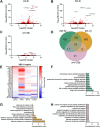
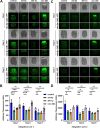
The capacity to deal with stress declines during the aging process, and preservation of cellular stress responses is critical to healthy aging. The unfolded protein response of the endoplasmic reticulum (UPRER) is one such conserved mechanism, which is critical for the maintenance of several major functions of the ER during stress, including protein folding and lipid metabolism. Hyperactivation of the UPRER by overexpression of the major transcription factor, xbp-1s, solely in neurons drives lifespan extension as neurons send a neurotransmitter-based signal to other tissue to activate UPRER in a non-autonomous fashion. Previous work identified serotonergic, dopaminergic, and tyraminergic neurons in this signaling paradigm. To further expand our understanding of the neural circuitry that underlies the non-autonomous signaling of ER stress, we activated UPRER solely in glutamatergic, octopaminergic, and GABAergic neurons in C. elegans and paired whole-body transcriptomic analysis with functional assays. We found that UPRER-induced signals from glutamatergic neurons increased expression of canonical protein homeostasis pathways and octopaminergic neurons promoted pathogen response pathways; while minor, statistically significant changes were observed in lipid metabolism-related genes with GABAergic UPRER activation. These findings provide further evidence for the distinct role neuronal subtypes play in driving the diverse response to ER stress.
Keywords: Aging; ER; neurons; stress.
Conflict of interest statement
Competing Financial Interests All authors of the manuscript declare that they have no competing interests.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
