Emergence of an Extensive Drug Resistant Citrobacter portucalensis Clinical Strain Harboring bla SFO-1, bla KPC-2, and bla NDM-1
- PMID: 38854780
- PMCID: PMC11162216
- DOI: 10.2147/IDR.S461118
Emergence of an Extensive Drug Resistant Citrobacter portucalensis Clinical Strain Harboring bla SFO-1, bla KPC-2, and bla NDM-1
Abstract
Background: To explore the plasmid characteristics and transfer mechanisms of an extensive drug resistant (XDR) clinical isolate, Citrobacter portucalensis L2724hy, co-producing bla SFO-1, bla NDM-1, and bla KPC-2.
Methods: Species confirmation of L2724hy was achieved through 16S rRNA sequencing and Average Nucleotide Identity (ANI) analysis. Antimicrobial susceptibility testing (AST) employed the agar dilution and micro broth dilution methods. Identification of resistance genes was carried out by PCR and whole-genome sequencing (WGS). Essential resistance gene locations were verified by S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and southern hybridization experiments. Subsequent WGS data analysis delved into drug resistance genes and plasmids.
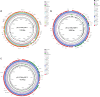
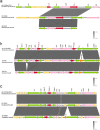
Results: The confirmation of the strain L2724hy as an extensive drug-resistant Citrobacter portucalensis, resistant to almost all antibiotics tested except polymyxin B and tigecycline, was achieved through 16S rRNA sequencing, ANI analysis and AST results. WGS and subsequent analysis revealed L2724hy carrying bla SFO-1, bla NDM-1, and bla KPC-2 on plasmids of various sizes. The uncommon ESBL gene bla SFO-1 coexists with the fosA3 gene on an IncFII plasmid, featuring the genetic environment IS26-fosA3-IS26-ampR-bla SFO-1-IS26. The bla NDM-1 was found on an IncX3 plasmid, coexisting with bla SHV-12, displaying the sequence IS5-IS3000-IS3000-Tn2-bla NDM-1-ble-trpF-dsbD-cutA-gros-groL, lacking ISAa125. The bla KPC-2 is located on an unclassified plasmid, exhibiting the sequence Tn2-tnpR-ISKpn27-bla KPC-2-ISKpn6-korC. Conjugation assays confirmed the transferability of both bla NDM-1 and bla KPC-2.
Conclusion: We discovered the coexistence of bla SFO-1, bla NDM-1, and bla KPC-2 in C. portucalensis for the first time, delving into plasmid characteristics and transfer mechanisms. Our finding highlights the importance of vigilant monitoring of drug-resistance genes and insertion elements in uncommon strains.
Keywords: Citrobacter spp; IncFII; XDR; blaKPC; blaNDM; blaSFO.
© 2024 Guo et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures


Similar articles
-
First documentation of a clinical multidrug-resistant Enterobacter chuandaensis ST2493 isolate co-harboring blaNDM-1 and two blaKPC-2 bearing plasmids.Sci Rep. 2024 Nov 5;14(1):26817. doi: 10.1038/s41598-024-78163-2. Sci Rep. 2024. PMID: 39500966 Free PMC article.
-
Coexistence of blaIMP-4, blaNDM-1 and blaOXA-1 in blaKPC-2-producing Citrobacter freundii of clinical origin in China.Front Microbiol. 2023 Jun 12;14:1074612. doi: 10.3389/fmicb.2023.1074612. eCollection 2023. Front Microbiol. 2023. PMID: 37378293 Free PMC article.
-
Genomic characterization of a multidrug-resistant Citrobacter portucalensis isolate co-harboring bla KPC-2 and bla NDM-1 on distinct plasmids.Front Microbiol. 2025 Jul 16;16:1633493. doi: 10.3389/fmicb.2025.1633493. eCollection 2025. Front Microbiol. 2025. PMID: 40740322 Free PMC article.
-
Emergence of Extensively Drug-Resistant ST170 Citrobacter portucalensis with Plasmids pK218-KPC, pK218-NDM, and pK218-SHV from a Tertiary Hospital, China.Microbiol Spectr. 2022 Oct 26;10(5):e0251022. doi: 10.1128/spectrum.02510-22. Epub 2022 Sep 26. Microbiol Spectr. 2022. PMID: 36154205 Free PMC article.
-
First Report of Coexistence of bla SFO-1 and bla NDM-1 β-Lactamase Genes as Well as Colistin Resistance Gene mcr-9 in a Transferrable Plasmid of a Clinical Isolate of Enterobacter hormaechei.Front Microbiol. 2021 Jun 18;12:676113. doi: 10.3389/fmicb.2021.676113. eCollection 2021. Front Microbiol. 2021. PMID: 34220761 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

