Senescence-targeted MicroRNA/Organoid composite hydrogel repair cartilage defect and prevention joint degeneration via improved chondrocyte homeostasis
- PMID: 38855061
- PMCID: PMC11157121
- DOI: 10.1016/j.bioactmat.2024.05.036
Senescence-targeted MicroRNA/Organoid composite hydrogel repair cartilage defect and prevention joint degeneration via improved chondrocyte homeostasis
Abstract
Introduction: Cartilage defect (CD) is a common complication in osteoarthritis (OA). Impairment of chondrogenesis and cellular senescence are considered as hallmarks of OA development and caused failure of cartilage repair in most clinical CD cases. Exploring markers for cellular senescence in CD patients might provide new perspectives for osteoarthritic CD patients. In the present study, we aim to explore senescent markers in CD patients with OA to fabricate a senescence-targeted SMSC organoid hydrogel for cartilage repair.
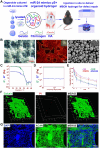
Methods: Clinical cartilage samples from cartilage defect patients were collected. Immunofluorescence staining of senescent markers and SA-β-Gal staining were used to detect the senescence state of SMSCs and chondrocytes in cartilage defect and OA patients. MicroRNA expression profiles of SMSC organoids and H2O2-treated SMSC organoids were analyzed and compared with high-throughput microRNA sequencing. Fluorescent in situ hybridization of miRNA were used to determine the expression level of miR-24 in SMSC organoids and cartilage samples. Interaction between miR-24 and its downstream target was analyzed via qRT-PCR, immunofluorescence and luciferase assay. Senescence-targeted miR-24 μS/SMSC organoid hydrogel (MSOH) was constructed for cartilage repair. Anti-senescence properties and chondrogenesis were determined in vitro for MSOH. Rats were used to evaluate the cartilage repair capacity of the MSOH hydrogel in vivo.
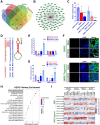
Results: In this study, we found Osteoarthritic cartilage defect patients demonstrated upregulated cellular senescence in joint cartilage. MicroRNA sequencing demonstrated senescence marker miR-24 was negatively associated with cartilage impairment and cellular senescence in osteoarthritic CD patients. Moreover, miR-24 mimics alleviates cellular senescence to promote chondrogenesis by targeting downstream TAOK1. Also, miR-24 downregulated TAOK1 expression and promoted chondrogenesis in SMSC organoids. Senescence-targeted miR-24 μS/SMSC organoid hydrogel (MSOH) was constructed and demonstrated superior chondrogenesis in vitro. Animal experiments demonstrated that MSOH hydrogel showed better cartilage repairing effects and better maintained joint function at 24 weeks with low intra-articular inflammatory response after transplantation in rat joint. Single-cell RNA-seq of generated cartilage indicated that implanted MSOH could affect chondrocyte homeostatic state and alter the chondrocyte cluster frequency by regulating cellular glycolysis and OXPHOS, impacting cell cycle and ferroptosis to alleviate cellular senescence and prevent joint degeneration.
Conclusion: Osteoarthritic cartilage defect patients demonstrated upregulated cellular senescence in joint cartilage. Senescence marker miR-24 was negatively associated with cartilage impairment in osteoarthritic CD patients. miR-24 attenuates chondrocytes senescence and promotes chondrogenesis in SMSC organoids through targeting TAOK1. Senescence-targeted miR-24 microsphere/SMSC organoid composite hydrogel could successfully repair cartilage defect in osteoarthritic microenvironment via enhanced miR-24/TAOK1 signaling pathway, suggesting MSOH might be a novel therapy for cartilage repair in osteoarthritic CD patients.
© 2024 The Authors.
Conflict of interest statement
The authors have declared that no competing interest exists.
Figures








Similar articles
-
Genetically inspired organoids prevent joint degeneration and alleviate chondrocyte senescence via Col11a1-HIF1α-mediated glycolysis-OXPHOS metabolism shift.Clin Transl Med. 2024 Feb;14(2):e1574. doi: 10.1002/ctm2.1574. Clin Transl Med. 2024. PMID: 38314968 Free PMC article.
-
[Effects of cartilage progenitor cells and microRNA-140 on repair of osteoarthritic cartilage injury].Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2019 May 15;33(5):650-658. doi: 10.7507/1002-1892.201806060. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2019. PMID: 31090363 Free PMC article. Review. Chinese.
-
Exosomes derived from miR-155-5p-overexpressing synovial mesenchymal stem cells prevent osteoarthritis via enhancing proliferation and migration, attenuating apoptosis, and modulating extracellular matrix secretion in chondrocytes.Cell Biol Toxicol. 2021 Feb;37(1):85-96. doi: 10.1007/s10565-020-09559-9. Epub 2020 Oct 25. Cell Biol Toxicol. 2021. PMID: 33099657
-
Asporin regulated by miR-26b-5p mediates chondrocyte senescence and exacerbates osteoarthritis progression via TGF-β1/Smad2 pathway.Rheumatology (Oxford). 2022 May 30;61(6):2631-2643. doi: 10.1093/rheumatology/keab725. Rheumatology (Oxford). 2022. PMID: 34559207
-
Preparation of hydrogel microsphere and its application in articular cartilage injury.Mater Today Bio. 2025 Mar 8;31:101641. doi: 10.1016/j.mtbio.2025.101641. eCollection 2025 Apr. Mater Today Bio. 2025. PMID: 40130039 Free PMC article. Review.
Cited by
-
Intelligent Manufacturing for Osteoarthritis Organoids.Cell Prolif. 2025 Jul;58(7):e70043. doi: 10.1111/cpr.70043. Epub 2025 Apr 26. Cell Prolif. 2025. PMID: 40285592 Free PMC article. Review.
-
Targeted Inhibition of cGAS/STING signaling induced by aberrant R-Loops in the nucleus pulposus to alleviate cellular senescence and intervertebral disc degeneration.J Nanobiotechnology. 2025 Jul 14;23(1):510. doi: 10.1186/s12951-025-03579-5. J Nanobiotechnology. 2025. PMID: 40660286 Free PMC article.
-
Skeletal organoids.Biomater Transl. 2024 Nov 15;5(4):390-410. doi: 10.12336/biomatertransl.2024.04.005. eCollection 2024. Biomater Transl. 2024. PMID: 39872931 Free PMC article. Review.
-
Regulation of ferroptosis in osteoarthritis and osteoarthritic chondrocytes by typical MicroRNAs in chondrocytes.Front Med (Lausanne). 2024 Nov 5;11:1478153. doi: 10.3389/fmed.2024.1478153. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39564502 Free PMC article. Review.
-
Future perspectives: advances in bone/cartilage organoid technology and clinical potential.Biomater Transl. 2024 Nov 15;5(4):425-443. doi: 10.12336/biomatertransl.2024.04.007. eCollection 2024. Biomater Transl. 2024. PMID: 39872930 Free PMC article. Review.
References
-
- Parker C., et al. Osteochondral allograft transplantation for knee cartilage and osteochondral defects: a review of indications, technique, rehabilitation, and outcomes. JBJS Rev. 2019;7(6):e7. - PubMed
-
- Ajay K., et al. High tibial osteotomies for the treatment of osteoarthritis of the knee. JBJS Rev. 2022;10(1) e21.00127. - PubMed
-
- Jason M J., Lindsay K., Michael B. Medial unicompartmental arthroplasty of the knee. J. Am. Acad. Orthop. Surg. 2019;27(5):166–176. - PubMed
LinkOut - more resources
Full Text Sources

