Single-cell analyses reveal evolution mimicry during the specification of breast cancer subtype
- PMID: 38855191
- PMCID: PMC11155410
- DOI: 10.7150/thno.96163
Single-cell analyses reveal evolution mimicry during the specification of breast cancer subtype
Abstract
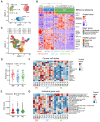
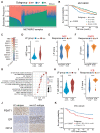
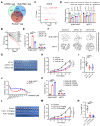
Background: The stem or progenitor antecedents confer developmental plasticity and unique cell identities to cancer cells via genetic and epigenetic programs. A comprehensive characterization and mapping of the cell-of-origin of breast cancer using novel technologies to unveil novel subtype-specific therapeutic targets is still absent. Methods: We integrated 195,144 high-quality cells from normal breast tissues and 406,501 high-quality cells from primary breast cancer samples to create a large-scale single-cell atlas of human normal and cancerous breasts. Potential heterogeneous origin of malignant cells was explored by contrasting cancer cells against reference normal epithelial cells. Multi-omics analyses and both in vitro and in vivo experiments were performed to screen and validate potential subtype-specific treatment targets. Novel biomarkers of identified immune and stromal cell subpopulations were validated by immunohistochemistry in our cohort. Results: Tumor stratification based on cancer cell-of-origin patterns correlated with clinical outcomes, genomic aberrations and diverse microenvironment constitutions. We found that the luminal progenitor (LP) subtype was robustly associated with poor prognosis, genomic instability and dysfunctional immune microenvironment. However, the LP subtype patients were sensitive to neoadjuvant chemotherapy (NAC), PARP inhibitors (PARPi) and immunotherapy. The LP subtype-specific target PLK1 was investigated by both in vitro and in vivo experiments. Besides, large-scale single-cell profiling of breast cancer inspired us to identify a range of clinically relevant immune and stromal cell subpopulations, including subsets of innate lymphoid cells (ILCs), macrophages and endothelial cells. Conclusion: The present single-cell study revealed the cellular repertoire and cell-of-origin patterns of breast cancer. Combining single-cell and bulk transcriptome data, we elucidated the evolution mimicry from normal to malignant subtypes and expounded the LP subtype with vital clinical implications. Novel immune and stromal cell subpopulations of breast cancer identified in our study could be potential therapeutic targets. Taken together, Our findings lay the foundation for the precise prognostic and therapeutic stratification of breast cancer.
Keywords: breast cancer; molecular subtype; single-cell RNA-seq; tumor cell-of-origin; tumor microenvironment.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




References
-
- Prat A, Perou CM. Mammary development meets cancer genomics. Nat Med. 2009;15:842–4. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

