A one health approach for monitoring antimicrobial resistance: developing a national freshwater pilot effort
- PMID: 38855419
- PMCID: PMC11157689
- DOI: 10.3389/frwa.2024.1359109
A one health approach for monitoring antimicrobial resistance: developing a national freshwater pilot effort
Abstract
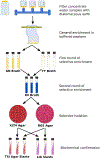
Antimicrobial resistance (AMR) is a world-wide public health threat that is projected to lead to 10 million annual deaths globally by 2050. The AMR public health issue has led to the development of action plans to combat AMR, including improved antimicrobial stewardship, development of new antimicrobials, and advanced monitoring. The National Antimicrobial Resistance Monitoring System (NARMS) led by the United States (U.S) Food and Drug Administration along with the U.S. Centers for Disease Control and U.S. Department of Agriculture has monitored antimicrobial resistant bacteria in retail meats, humans, and food animals since the mid 1990's. NARMS is currently exploring an integrated One Health monitoring model recognizing that human, animal, plant, and environmental systems are linked to public health. Since 2020, the U.S. Environmental Protection Agency has led an interagency NARMS environmental working group (EWG) to implement a surface water AMR monitoring program (SWAM) at watershed and national scales. The NARMS EWG divided the development of the environmental monitoring effort into five areas: (i) defining objectives and questions, (ii) designing study/sampling design, (iii) selecting AMR indicators, (iv) establishing analytical methods, and (v) developing data management/analytics/metadata plans. For each of these areas, the consensus among the scientific community and literature was reviewed and carefully considered prior to the development of this environmental monitoring program. The data produced from the SWAM effort will help develop robust surface water monitoring programs with the goal of assessing public health risks associated with AMR pathogens in surface water (e.g., recreational water exposures), provide a comprehensive picture of how resistant strains are related spatially and temporally within a watershed, and help assess how anthropogenic drivers and intervention strategies impact the transmission of AMR within human, animal, and environmental systems.
Keywords: antimicrobial resistance; environment; freshwater; human health; monitoring; one health; surface waters.
Conflict of interest statement
Conflict of interest The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
National Antimicrobial Resistance Monitoring System: Two Decades of Advancing Public Health Through Integrated Surveillance of Antimicrobial Resistance.Foodborne Pathog Dis. 2017 Oct;14(10):545-557. doi: 10.1089/fpd.2017.2283. Epub 2017 Aug 9. Foodborne Pathog Dis. 2017. PMID: 28792800 Free PMC article. Review.
-
A National Antimicrobial Resistance Monitoring System Survey of Antimicrobial-Resistant Foodborne Bacteria Isolated from Retail Veal in the United States.J Food Prot. 2021 Oct 1;84(10):1749-1759. doi: 10.4315/JFP-21-005. J Food Prot. 2021. PMID: 34015113 Free PMC article.
-
Retracted: Antibiotic Resistance in Salmonella Enteritidis Isolates Recovered from Chicken, Chicken Breast, and Humans Through National Antimicrobial Resistance Monitoring System Between 1996 and 2014.Foodborne Pathog Dis. 2018 Oct 3. doi: 10.1089/fpd.2017.2402. Online ahead of print. Foodborne Pathog Dis. 2018. Retraction in: Foodborne Pathog Dis. 2018 Oct;15(10):669. doi: 10.1089/fpd.2017.2402.retract. PMID: 29927626 Retracted.
-
Antimicrobial Resistance and Environmental Health: A Water Stewardship Framework for Global and National Action.Antibiotics (Basel). 2022 Jan 5;11(1):63. doi: 10.3390/antibiotics11010063. Antibiotics (Basel). 2022. PMID: 35052940 Free PMC article.
-
Averting the AMR crisis: What are the avenues for policy action for countries in Europe? [Internet].Copenhagen (Denmark): European Observatory on Health Systems and Policies; 2019. Copenhagen (Denmark): European Observatory on Health Systems and Policies; 2019. PMID: 31287637 Free Books & Documents. Review.
Cited by
-
Population Ecology-Quantitative Microbial Risk Assessment (QMRA) Model for Antibiotic-Resistant and Susceptible E. coli in Recreational Water.Environ Sci Technol. 2025 Mar 11;59(9):4266-4281. doi: 10.1021/acs.est.4c07248. Epub 2025 Feb 26. Environ Sci Technol. 2025. PMID: 40008406
-
Comprehensive regional study of ESBL Escherichia coli: genomic insights into antimicrobial resistance and inter-source dissemination of ESBL genes.Front Microbiol. 2025 Jun 10;16:1595652. doi: 10.3389/fmicb.2025.1595652. eCollection 2025. Front Microbiol. 2025. PMID: 40556893 Free PMC article.
-
Bioinspired Nanoplatforms: Polydopamine and Exosomes for Targeted Antimicrobial Therapy.Polymers (Basel). 2025 Jun 16;17(12):1670. doi: 10.3390/polym17121670. Polymers (Basel). 2025. PMID: 40574198 Free PMC article. Review.
-
Evaluating Quantitative Metagenomics for Environmental Monitoring of Antibiotic Resistance and Establishing Detection Limits.Environ Sci Technol. 2025 Apr 1;59(12):6192-6202. doi: 10.1021/acs.est.4c08284. Epub 2025 Mar 18. Environ Sci Technol. 2025. PMID: 40100955 Free PMC article.
-
Agriculturally Sourced Multidrug-Resistant Escherichia coli for Use as Control Strains.Pathogens. 2025 Apr 25;14(5):417. doi: 10.3390/pathogens14050417. Pathogens. 2025. PMID: 40430738 Free PMC article.
References
-
- Alakomi H-L, and Saarela M (2009). Salmonella importance and current status of detection and surveillance methods. Qual. Assur. Safety Crops Food 1, 142–152. doi: 10.1111/j.1757-837X.2009.00032.x - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
