Deep learning based characterization of human organoids using optical coherence tomography
- PMID: 38855657
- PMCID: PMC11161340
- DOI: 10.1364/BOE.515781
Deep learning based characterization of human organoids using optical coherence tomography
Abstract
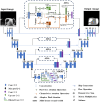
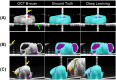
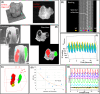
Organoids, derived from human induced pluripotent stem cells (hiPSCs), are intricate three-dimensional in vitro structures that mimic many key aspects of the complex morphology and functions of in vivo organs such as the retina and heart. Traditional histological methods, while crucial, often fall short in analyzing these dynamic structures due to their inherently static and destructive nature. In this study, we leveraged the capabilities of optical coherence tomography (OCT) for rapid, non-invasive imaging of both retinal, cerebral, and cardiac organoids. Complementing this, we developed a sophisticated deep learning approach to automatically segment the organoid tissues and their internal structures, such as hollows and chambers. Utilizing this advanced imaging and analysis platform, we quantitatively assessed critical parameters, including size, area, volume, and cardiac beating, offering a comprehensive live characterization and classification of the organoids. These findings provide profound insights into the differentiation and developmental processes of organoids, positioning quantitative OCT imaging as a potentially transformative tool for future organoid research.
© 2024 Optica Publishing Group.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Update of
- doi: 10.1364/opticaopen.24790584.
References
-
- Shoji J. Y., Davis R. P., Mummery C. L., et al. , “Global meta-analysis of organoid and organ-on-chip research,” Adv. Healthcare Mater., 2301067 (2023). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
