LexA, an SOS response repressor, activates TGase synthesis in Streptomyces mobaraensis
- PMID: 38855760
- PMCID: PMC11157053
- DOI: 10.3389/fmicb.2024.1397314
LexA, an SOS response repressor, activates TGase synthesis in Streptomyces mobaraensis
Abstract
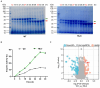
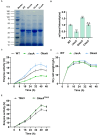
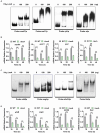
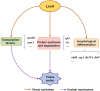
Transglutaminase (EC 2.3.2.13, TGase), an enzyme that catalyzes the formation of covalent cross-links between protein or peptide molecules, plays a critical role in commercial food processing, medicine, and textiles. TGase from Streptomyces is the sole commercial enzyme preparation for cross-linking proteins. In this study, we revealed that the SOS response repressor protein LexA in Streptomyces mobaraensis not only triggers morphological development but also enhances TGase synthesis. The absence of lexA significantly diminished TGase production and sporulation. Although LexA does not bind directly to the promoter region of the TGase gene, it indirectly stimulates transcription of the tga gene, which encodes TGase. Furthermore, LexA directly enhances the expression of genes associated with protein synthesis and transcription factors, thus favorably influencing TGase synthesis at both the transcriptional and posttranscriptional levels. Moreover, LexA activates four crucial genes involved in morphological differentiation, promoting spore maturation. Overall, our findings suggest that LexA plays a dual role as a master regulator of the SOS response and a significant contributor to TGase regulation and certain aspects of secondary metabolism, offering insights into the cellular functions of LexA and facilitating the strategic engineering of TGase overproducers.
Keywords: LexA; TGase; regulation; streptomyces; transcription factor.
Copyright © 2024 Shi, Yan, Yuan, Li, Liu, Li, Yu, Li, Zhu and Wang.
Conflict of interest statement
FY and GL were employed by Jiangsu Yiming Biological Technology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer LB declared a past co-authorship with the author WW to the handling editor.
Figures


Similar articles
-
Identification of multiple regulatory genes involved in TGase production in Streptomyces mobaraensis DSM 40587.Eng Microbiol. 2023 Jun 10;3(4):100098. doi: 10.1016/j.engmic.2023.100098. eCollection 2023 Dec. Eng Microbiol. 2023. PMID: 39628909 Free PMC article.
-
Enhanced Production of Transglutaminase in Streptomyces mobaraensis through Random Mutagenesis and Site-Directed Genetic Modification.J Agric Food Chem. 2021 Mar 17;69(10):3144-3153. doi: 10.1021/acs.jafc.1c00645. Epub 2021 Mar 2. J Agric Food Chem. 2021. PMID: 33651593
-
Genome-Wide Identification of the LexA-Mediated DNA Damage Response in Streptomyces venezuelae.J Bacteriol. 2022 Aug 16;204(8):e0010822. doi: 10.1128/jb.00108-22. Epub 2022 Jul 13. J Bacteriol. 2022. PMID: 35862789 Free PMC article.
-
Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon.Mol Microbiol. 2006 Dec;62(5):1228-38. doi: 10.1111/j.1365-2958.2006.05444.x. Epub 2006 Oct 17. Mol Microbiol. 2006. PMID: 17042786 Review.
-
The Use and Abuse of LexA by Mobile Genetic Elements.Trends Microbiol. 2016 May;24(5):391-401. doi: 10.1016/j.tim.2016.02.009. Epub 2016 Mar 9. Trends Microbiol. 2016. PMID: 26970840 Review.
References
-
- Bellio P., Mancini A., Di Pietro L., Cracchiolo S., Franceschini N., Reale S., et al. . (2020). Inhibition of the transcriptional repressor LexA: withstanding drug resistance by inhibiting the bacterial mechanisms of adaptation to antimicrobials. Life Sci. 241:117116. doi: 10.1016/j.lfs.2019.117116, PMID: - DOI - PubMed
-
- Ceresino E. B., de Melo R. R., Kuktaite R., Hedenqvist M. S., Zucchi T. D., Johansson E., et al. . (2018). Transglutaminase from newly isolated Streptomyces sp. CBMAI 1617: production optimization, characterization and evaluation in wheat protein and dough systems. Food Chem. 241, 403–410. doi: 10.1016/j.foodchem.2017.09.010, PMID: - DOI - PubMed
-
- Chen K., Zhang D., Liu S., Wang N. S., Wang M., Du G., et al. . (2013). Improvement of transglutaminase production by extending differentiation phase of Streptomyces hygroscopicus: mechanism and application. Appl. Microbiol. Biotechnol. 97, 7711–7719. doi: 10.1007/s00253-012-4614-y, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources

